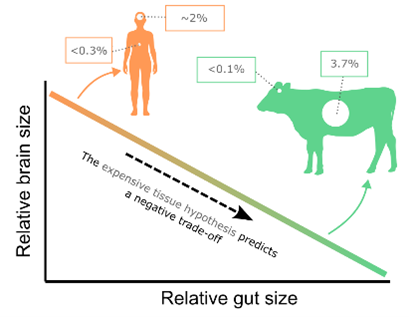
This PhD project will provide new insights into how evolutionary trade-offs have shaped macro-evolutionary diversity, with a particular emphasis on the “expensive tissue hypothesis” (ETH). The expensive tissue hypothesis originally proposed that animals – such as our own big-brained species – could only evolve such large brains by reducing energetic investment in other expensive tissues such as the gut. It has since been expanded to propose that in the evolutionary arms race, various costly organs such as the brain, heart, digestive system, and liver, are all interlinked in a series of trade-offs. Increasing the size of one organ is likely to necessitate the reduction of another or an increase in overall metabolic rate to accommodate increased energetic demands. This would result in a negative trade-off between the size of expensive organs across species (Fig. 1). This idea has been very influential yet highly controversial, with variable support across different tissues and taxa.
Despite its widespread influence, the last organ data compilation used to test such ideas dates to 1940 and has since been widely cited. Since then, small bespoke datasets have been compiled for specific taxa or tissues, making it difficult to draw generalized conclusions. This project will address this gap by compiling a comprehensive dataset that includes organ size data spanning all vertebrate species and from all available sources including wildlife institutions, data repositories and primary literature. This new dataset will enable a robust, macro-evolutionary test of the ETH and its causes, exploring whether proposed trade-offs hold true across a broad range of species and evolutionary contexts.
Another major aspect of the project will be to harness the power of the data collected spanning multiple individuals within a single species to examine intraspecific variation and trade-offs in organ sizes. This dual approach – combining broad, cross-species comparisons with detailed within-species analyses sets this PhD apart and facilitates a nuanced understanding of how these trade-offs might evolve and function at different evolutionary scales. This will give rise to exciting new insight into how macroevolutionary patterns can arise from within-species variation and adaptation.
Figure 1: The Expensive Tissue Hypothesis.
This project is not suitable for CASE funding
Each host has a slightly different application process.
Find out how to apply for this studentship.
All applications must include the CENTA application form.
Choose your application route
The student will quantify variation in organ sizes at multiple scales (both within and across vertebrate species). Using phylogenetic comparative methods (e.g. “variable rates” models, approximate Bayesian computation, the fabric model), they will characterize the coevolution of major organs. The approaches allow simultaneous estimation of the location and magnitude of historical accelerations (or decelerations) in evolutionary rate, relationships amongst organs, as well as underlying phylogenetic signal. These rates will then be used to test underlying relationships and quantify historical adaptation. The project will also use methods for studying within- and between- species variation and how they interact within a single model: Bayesian phylogenetic linear mixed models implemented in multiple software packages. For all aspects of the project, various programs and R packages for phylogenetic comparative analysis will be utilized. This includes a package that is currently under development (BayesTraitR) that the student may also be involved in depending on their interests.
DRs will be awarded CENTA Training Credits (CTCs) for participation in CENTA-provided and ‘free choice’ external training. One CTC can be earned per 3 hours training, and DRs must accrue 100 CTCs across the three and a half years of their PhD.
As a part of direct experience and integration into a productive research group, the student will be trained in phylogenetic comparative methods, data collection, management, and reproducible workflows, alongside computational analysis and coding skills in R and Python. Further to this, the student will receive core training as a member of the postgraduate community at the Open University, including access to a wide range of research development programme skills – all of which are mapped to the VITAE Researcher Development Framework (RDF). Many of these will be highly desirable transferable skills including data handling, communication, project management, leadership and wellbeing.
Chris Venditti (Co-Supervisor) is internationally recognized in developing and applying phylogenetic comparative analyses. He has pioneered statistical frameworks for testing evolutionary hypotheses across large datasets, making his input invaluable. His involvement will provide the student with access to unparalleled training opportunities and enhance the projects impact.
Alice Johnston (Co-Supervisor) brings unique expertise in studying the energetics of diverse organisms, individuals, and use of agent-based modelling. Her research integrates detailed data with computational simulations and provides powerful tools to link individual level processes with large-scale evolutionary patterns. Her involvement will provide students the opportunity to engage in a diverse breadth of specialist methodologies.
Year 1: The first year of the project will be focussed on the compilation and standardisation of organ size data across vertebrates spanning the literature, museums, and institutional sources. The student will be trained in advanced comparative methods with the aim of conducting exploratory analyses using existing data: exploring organ size distributions and scaling relationships. There are two major objectives in this first year.
Year 2: The second year of the project will focus on macro-evolutionary change, using phylogenetic comparative analysis to test for trade-offs amongst organs. This will involve the identification of rate shifts, correlations, and historical adaptations across the vertebrate phylogeny. There are two major objectives for the second year.
Year 3: The final year of the project will be dedicated to incorporating data spanning individuals to examine intraspecific variation. This will involve Bayesian phylogenetic linear mixed models to integrate the study of both within- and between- species patterns. There are three major objectives of this year.
The expensive tissue hypothesis:
Comparative analysis and the types of questions we can answer:
The last comprehensive database of organ size:
Students should have a background in biology or zoology, and an interest in evolution or ecology. The PhD student will join a vibrant postgraduate community at the Open University within the Faculty of Science, Technology, Engineering and Mathematics (STEM).
If you are interested in this project, we recommend you get in touch with the primary contact, Dr Joanna Baker ([email protected]) for an informal discussion and advice. For any procedural or administrative questions please email [email protected].
To apply to this project:
Applications must be submitted by 23:59 GMT on Wednesday 7th January 2026.