The world’s forests are under pressure from combined land use and climate change, with major consequences for biodiversity and the essential ecosystem services they provide. Critical ecological functions include soil carbon storage, nutrient cycling and regulation of greenhouse gases (GHGs), which are primarily driven by soil microbes. Large-scale forest restoration initiatives aim to address the biodiversity and climate change crises globally (e.g. Bonn Challenge) as well as in the UK, where extensive tree planting programmes form a central part of national Net Zero and Nature Recovery policy. Understanding the role that belowground biodiversity plays in forest recovery is key for successful rehabilitation of forest ecosystems and functions, but initiatives are often hindered by limited knowledge of plant-soil interactions during ecological restoration. Moreover, ensuring resilience of UK forests to future climate change requires insights into how native tree populations and associated belowground communities adapt to, and interact with, their environment.
Genetic variation among and within species provides the raw material for adaptation. Most studies assessing tree genetic diversity have focussed on aboveground traits, while less is known about belowground traits and links to soil microbes. Some evidence suggests that within species genetic variation in trees can influence the composition of soil microbial communities, which may in turn affect tree establishment and nutrient cycling. Furthermore, there is uncertainty around the spatial and temporal scale of these processes, as soil microbial communities are strongly affected by their environment and can vary significantly over centimetres to metres, seasonally and over tree life cycles.
This project will examine links between tree intraspecific variation, soil microbial communities and biogeochemical (carbon and nutrient) cycling to support effective restoration and resilience in Scottish Caledonian pinewoods – the only UK habitat with continuous links back to post-glacial vegetation. Research will focus on Scots pine, a keystone species of Caledonian forest with global ecological and economic importance and the 2nd most planted species in the UK. This studentship suits an individual interested in evaluating adaptive traits and soil functions in field and laboratory settings, and understanding how this acquired knowledge can be translated into policy for British woodlands.
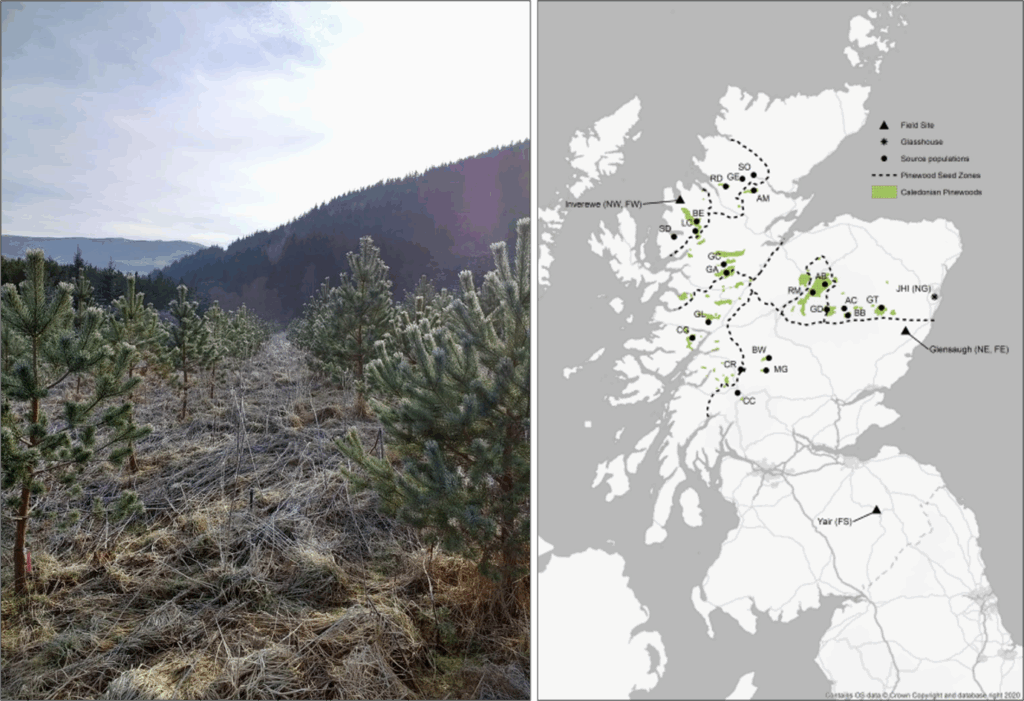
Figure 1: (Left) Young Scots pine trees of different genotypes growing in a common garden environment at one of the established field trials in Scotland. (Right) Map of the 21 Caledonian pinewood populations across Scotland from which seeds were collected (black circles), and locations of the three established trial sites (black triangles) – figure adapted from Beaton et al. (2022).
This project is a CENTA Flagship Project.
This project is suitable for CASE funding
Each host has a slightly different application process.
Find out how to apply for this studentship.
All applications must include the CENTA application form.
Choose your application route
The student will gain access to existing long-term common garden experiments and seed banks established by UKCEH and Forest Research to assess genetic diversity in native Scots pine. These comprise trees from 21 Scottish populations which have been extensively characterised for genotypic and aboveground phenotypic traits since planting in 2012. Activities will utilise these invaluable resources to link tree genetic variation, soil microbial communities and biogeochemical cycling by combining (1) field survey of soil and belowground tree traits over varying spatiotemporal scales, (2) in-situ measurement of soil carbon/nutrient cycling (e.g. GHG fluxes) and (3) pot/laboratory experiments to test tree genetic control on soil microbial assembly and processes, including climate change impacts (warming and drought). Microbial communities and functions will be evaluated using advanced soil eDNA metabarcoding and state-of-the-art isotopic techniques. An additional unique opportunity is to study tree-soil interactions during early establishment as new trials will be established in 2026.
DRs will be awarded CENTA Training Credits (CTCs) for participation in CENTA-provided and ‘free choice’ external training. One CTC can be earned per 3 hours training, and DRs must accrue 100 CTCs across the three and a half years of their PhD.
This is an exciting opportunity to work with an experienced team who combine expertise in conservation genetics, molecular ecology and plant-soil biogeochemistry. Several team members have successful track records of PhD supervision. Training will be provided in soil, plant and biogeochemical surveys/analysis (including soil-atmosphere GHG exchange), eDNA metabarcoding (DNA extraction, PCR, processing/analysing molecular and environmental data) and design/execution of laboratory experiments (including advanced isotopic techniques and controlled-environment facilities). The research institutions involved offer the valuable opportunity to gain experience in both fundamental and applied research settings. The student will have access to additional training provided through the pan-European EVOLTREE network.
Forest Research is the internationally–renowned research agency of the Forestry Commission and Britain’s principal forestry and tree-related research organisation. It provides science, evidence, data and services for sustainable forestry, supporting improvement and delivery of UK policy. The Royal Botanic Gardens, Kew is a world-leading plant and fungal research institution. Doctoral students aligned with Kew have access to Kew’s collections and facilities, including the herbarium, fungarium and living collections. They also join Kew’s community of >130 PhD researchers, with opportunities to engage with wider public programmes, advocacy and education work, an annual PhD symposium, regular student competitions, talks and research activities.
Year 1: Design and carry out soil and biogeochemical surveys of established common garden experiments to be repeated seasonally over an annual cycle.
Year 2: Complete soil and biogeochemical surveys of established field trials, DNA sequencing of soil samples and analysis. Establish pot experiments using seedbank material and contrasting soil types, to assess plasticity in root traits and rhizosphere soil microbial communities in a controlled environment.
Year 3: Complete pot experiments, design and conduct controlled laboratory incubations using rhizosphere soil of different Scots pine genotypes to assess microbial and biogeochemical responses to future climate change (experimental warming and drought).
Beaton, J., Perry, A., Cottrell, J., Iason, G., Stockan, J. & Cavers, S. (2022) ‘Phenotypic trait variation in a long-term multisite common garden experiment of Scots pine in Scotland’, Scientific Data, 9, 671.
Champion, A., Bazzicalupo, A., Heuertz, M., & Gargiulo, R. (In prep.) ‘Conservation genetics in ectomycorrhizal fungi: estimating the effective population size (Ne)’.
Downie, J., Silvertown, J., Cavers, S. & Ennos, R. (2020) ‘Heritable genetic variation but no local adaptation in a pine-ectomycorrhizal interaction’, Mycorrhiza, 30, pp. 185-195.
Downie, J., Taylor, A. F. S., Iason, G., Moore, B., Silvertown, J., Cavers, S. & Ennos, R. (2021) ‘Location, but not defensive genotype, determines ectomycorrhizal community composition in Scots pine (Pinus sylvestris L.) seedlings’, Ecology and Evolution, 11, pp. 4826-4842.
Gargiulo, R., Budde, K. B., & Heuertz, M. (2025) ‘Mind the lag: understanding genetic extinction debt for conservation’, TREE, 40, pp. 228-237.
Hazard, C. & Johnson, D. (2018) ‘Does genotypic and species diversity of mycorrhizal plants and fungi affect ecosystem function?’, New Phytologist, 220, pp. 1122-1128.
Korkama, T., Pakkanen, A. & Pennanen, T. (2006) ‘Ectomycorrhizal community structure varies among Norway spruce (Picea abies) clones’, New Phytologist, 171, pp. 815-824.
Patterson, A., Flores-Rentería, L., Whipple, A., Whitham, T. & Gehring, C. (2019) ‘Common garden experiments disentangle plant genetic and environmental contributions to ectomycorrhizal fungal community structure’, New Phytologist, 221, pp. 493-502.
Salmela, M. J., Velmala, S. M. & Pennanen, T. (2020) ‘Seedling traits from root to shoot exhibit genetic diversity and distinct responses to environmental heterogeneity within a tree population’, Oikos, 129, pp. 544-558.
Schönrogge, K., Gibbs, M., Oliver, A., Cavers, S., Gweon, H. S., Ennos, R. A., Cottrell, J., Iason, G. R. & Taylor, J. (2022) ‘Environmental factors and host genetic variation shape the fungal endophyte communities within needles of Scots pine (Pinus sylvestris)’, Fungal Ecology, 57-58, 101162.
Thomas, E., Jalonen, R., Loo, J., Boshier, D., Gallo, L., Cavers, S., Bordács, S., Smith, P. & Bozzano, M. (2014) ‘Genetic considerations in ecosystem restoration using native tree species’, Forest Ecology and Management, 333, pp. 66-75.
Velmala, S. M., Rajala, T., Haapanen, M., Taylor, A. F. S. & Pennanen, T. (2013) ‘Genetic host-tree effects on the ectomycorrhizal community and root characteristics of Norway spruce’, Mycorrhiza, 23, pp. 21-33.
For more information about this project please contact Sam Robinson at UKCEH Lancaster: [email protected]
Additional information about the European EVOLTREE (EVOLution of TREEs as drivers of terrestrial biodiversity) Research Network can be found on the website: https://www.evoltree.eu/
The successful applicant would be registered at the University of Warwick.
To apply to this project:
Applications must be submitted by 23:59 GMT on Wednesday 7th January 2026.