The plant microbiome, particularly in the phyllosphere (aboveground surfaces) and rhizosphere (rootassociated zone), plays a vital role in plant health, nutrient cycling, and disease resistance [1]. However, little is known about the ecological and evolutionary pressures shaping microbial community dynamics in these environments [2]. Bacteriophages, viruses that infect bacteria, are increasingly recognised as key drivers of microbial diversity and evolution, yet their influence in plantassociated environments remains poorly understood [3].
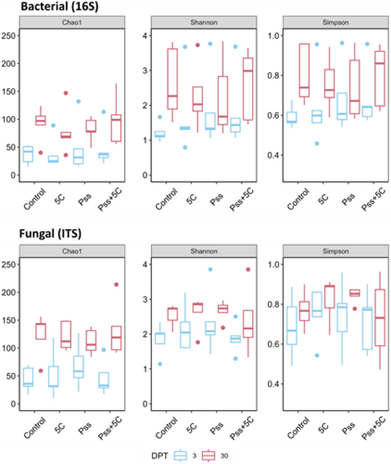
Our recent study showed that applying a phage cocktail to cherry leaves and shoots did not significantly impact microbial diversity (Figure 1) [4]. However, it remains unclear how naturally occurring phages shape microbial community structure and function in planta [5].
This project aims to investigate how natural phage populations influence microbial community composition, diversity, and function in the phyllosphere and rhizosphere. Using metagenomic and ampliconbased microbiome sequencing, we will characterise bacterial and viral communities across multiple crop species and environmental conditions [6]. Evolutionary experiments in controlled microcosms will be used to track phage–bacteria coevolution over time [7]. In parallel, metabolomic and transcriptomics will identify bacterial and viral genetic factors that mediate phage susceptibility and resistance in plant pathogenic bacteria, focusing on phage receptors and antiviral defence systems [8].
This research will provide new insights into the ecological roles of phages in plant environments and the evolutionary mechanisms that maintain microbial diversity [9]. The findings will inform sustainable plant microbiome management strategies and contribute to a broader understanding of viral ecology, aligning with the NERC remit on biodiversity, microbial resilience, and ecosystem function [10].
Figure 1. Alpha diversity indices of bacterial and fungal communities from cherry leaves sampled 3 and 30 days after treatment (DPT). The treatments were Pseudomonas syringae pv. syringae 9097, phage cocktail 5C, Pss combined with phage cocktail 5C, and a control. Diversity was evaluated using three indices: Chao1, estimating the total number of species per sample; Shannon’s index, reflecting community diversity (higher values indicate greater diversity); and Simpson’s index, reflecting community evenness (higher values indicate a more even community). Data adapted from Rabiey et al., 2025 [4].
This project is not suitable for CASE funding
Each host has a slightly different application process.
Find out how to apply for this studentship.
All applications must include the CENTA application form.
Choose your application route
DRs will be awarded CENTA Training Credits (CTCs) for participation in CENTA-provided and ‘free choice’ external training. One CTC can be earned per 3 hours training, and DRs must accrue 100 CTCs across the three and a half years of their PhD.
The student will receive specialist training for this multidisciplinary project, encompassing fieldwork, microbiology, metabolomics, transcriptomic, genomics, bioinformatic and data management and statistical analysis and interpretation of large and complex data sets.
The student will be supported to develop these skills within the SLS at Warwick, allowing the student to excel in these key aspects of data acquisition, analysis and dissemination and to build important networks.
The supervisory team is multi-disciplinary and highly experienced, based in excellent, well-equipped institution, and will provide comprehensive support for the student across all aspects of the project. The student will also have access to bioinformatic training for sequence analysis.
Not applicable.
We are not carrying out experiments on organisms that necessitate ethical or other types of approvals or licenses, other than necessary APHA compliance. There is no genetic or biological risk, as all bacteria are sourced directly from the UK natural environment. There are no plans to release genetically modified organisms during this project. This work is approved by the UoW under existing genetic manipulation and biological safety committee project reference 456.
For any enquiries related to this project please contact Mojgan Rabiey, [email protected].
To apply to this project:
Applications must be submitted by 23:59 GMT on Wednesday 7th January 2026.