Environmental insults such as pollution can cause transgenerational negative health outcomes in wild animal populations. Of particular concern are freshwater ecosystems, where 85% of species have experienced a decline since 1970 (WWF Living Planet Report 2024). Whilst mitigation strategies and policies are being put in place to reduce the impact of pollution, the consequences across generations have largely been ignored.
Epigenetic mechanisms, such as DNA methylation, can act as the causative agents, driving transgenerational health outcomes and/or act as a biomarker which can predict transgenerational health (Fig.1). Current research on pollution-induced epigenetic inheritance has focused on lab model systems – with limited applicability to “real-world” scenarios. This project will create a significant leap in this field by combining molecular epigenetics with natural population studies to understand the transgenerational and evolutionary impacts of pollution exposure.
Specifically, this PhD will examine DNA methylation differences between wild populations of the waterflea, Daphnia magna, from high and low pollution environments. Daphnia will be reared for multiple generations in the lab to determine if there are heritable DNA methylation marks associated with historic pollution exposure and if these marks can predict the health of future generations. This will provide evidence for transgenerational effects of pollution exposure (or lack thereof) which could feed into regulatory policy.
Questions this project will address:
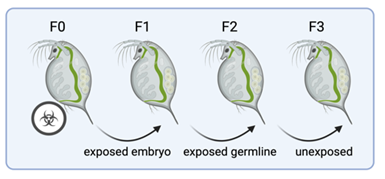
Figure 1: transgenerational inheritance is defined by the observation of an effect in the first unexposed generation – i.e. the first generation not exposed through the somatic tissue or the germline.
This project is not suitable for CASE funding
Each host has a slightly different application process.
Find out how to apply for this studentship.
All applications must include the CENTA application form.
Choose your application route
Fieldwork: Fieldwork will be carried out to sample wild populations of Daphnia magna from high and low pollution lakes.
Experimental evolution: Sampled Daphnia will be used to generate new clonal lines within the lab and examine their responses to novel pollution events – determining how much of the epigenome is stable and how much is driven by the environment.
Molecular lab work: DNA will be extracted from control and exposed samples and sequenced in-house using Nanopore technology. This will give both the genome and epigenome for all samples.
Bioinformatics: Genetic and epigenetic data will be analysed using custom pipelines to explore population genetics, population epigenetics and transgenerational epigenetics. Specifically, machine learning pipelines will be developed to predict pollution background from epigenomic data (i.e. generate epigenetic biomarkers).
DRs will be awarded CENTA Training Credits (CTCs) for participation in CENTA-provided and ‘free choice’ external training. One CTC can be earned per 3 hours training, and DRs must accrue 100 CTCs across the three and a half years of their PhD.
You will receive training in all aspects of the project. The UoL supervisor is an expert in ecological epigenetics and will provide in-house training for the molecular wet-lab work and the bioinformatic analysis of data, whilst the UoB supervisor is an expert in ecotoxicology and experimental evolution and will provide training through the state-of-the-art Daphnia Facility at UoB.
You will also be a member of the Centre for Environmental Health and Sustainability at UoL and of the Centre for Environmental Research and Justice at UoB. These groups offer a welcoming and positive research environment with opportunities for cross-disciplinary learning.
Year 1: Sample wild populations from multiple high and low pollution lakes; establish clonal lineages in the lab; carry out Nanopore sequencing on different clonal lines; use bioinformatic analyses to determine population-level genetic/epigenetic signatures of pollution.
Year 2: Expose clonal lineages to novel pollutants; examine if there is an evolved epigenetic response via Nanopore sequencing and bioinformatic analysis.
Year 3: Carry out transgenerational experiments to determine if phenotypic changes induced by pollution are heritable; generate machine learning pipelines to determine if historical epigenetic signatures can predict future population phenotypic outcomes.
Chaturvedi et al. (2023). The hologenome of Daphnia magna reveals possible DNA methylation and microbiome-mediated evolution of the host genome. Nucleic Acids Research. 51:18, 9785-9803.
Harney et al. (2022). Pollution induced epigenetic effects that are stably transmitted across multiple generations. Evolution Letters. 6:2, 118-135.
Venney et al. (2023). The evolutionary complexities of DNA methylation in animals: from plasticity to genetic evolution. Genome Biology and Evolution. 15:12, evad216.
Project contact details: [email protected]
To apply to this project:
Applications must be submitted by 23:59 GMT on Wednesday 7th January 2026.