Fleshy-fruited Myrtaceae represent a major lineage of tropical woody plants, comprising nearly half of the family’s ~6,000 species. These fruits are considered a key evolutionary innovation, linked to increased diversification rates and successful dispersal. This project explores the ecological and evolutionary implications of fleshy fruit evolution in Myrtaceae, focusing on two hyperdiverse tribes: Myrteae and Syzygieae. Using a combination of trait data, phylogenies, and environmental variables, the project aims to understand how fruit and seed traits shape niche dynamics, dispersal ability, and vulnerability to climate change. A major innovation of this work is the integration of AI tools to accelerate trait data acquisition across large, digitized collections and textual sources, unlocking new possibilities for trait-based research.
This project will leverage artificial intelligence to enhance both the efficiency and scope of trait data acquisition. Computer vision and deep learning models will be applied to digitized herbarium collections and field photographs to automatically extract fruit and seed traits, while large language models such as ChatGPT and Gemini will mine botanical literature and databases for complementary information. A key challenge is the integration of image-based and text-derived datasets, which differ in scale, structure, and accuracy. Addressing this requires robust data fusion and validation strategies. By combining these AI-derived datasets with phylogenies and environmental variables, the project will establish a framework to rapidly explore trait evolution, predict dispersal potential, and assess vulnerability to climate change. This project offers applicants the opportunity to work at the intersection of biodiversity science and cutting-edge AI, making it an exciting and impactful research topic.
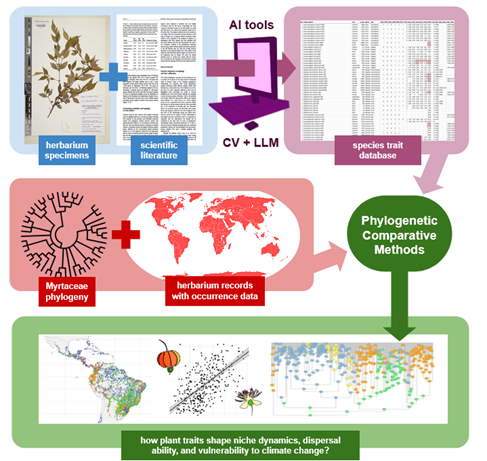
Figure 1: This image shows the use of computer vision and large language models to extract plant traits and investigate how these traits shape niche dynamics, dispersal ability, and vulnerability to climate change.
This project is not suitable for CASE funding
Each host has a slightly different application process.
Find out how to apply for this studentship.
All applications must include the CENTA application form.
Choose your application route
The project integrates phylogenetic comparative methods, ecological niche modeling, and machine learning for large-scale trait data acquisition. For Myrteae, a comprehensive fruit and seed trait database (~1,000 species) is being compiled through herbarium measurements and literature review. For Syzygieae, the project will deploy computer vision models (e.g., Mask R-CNN, YOLO) to extract trait data from herbarium specimen images and citizen science platforms. These models will be paired with large language models (LLMs) to interpret contextual information from specimen labels and literature. Phylogenetic frameworks for both tribes (with ~700 and ~300 tips, respectively) will support comparative analyses testing associations between fruit traits, environmental transitions, and range size. Finally, models of dispersal and colonization will assess the functional role of specific traits in insular and continental contexts under current and future climate scenarios.
DRs will be awarded CENTA Training Credits (CTCs) for participation in CENTA-provided and ‘free choice’ external training. One CTC can be earned per 3 hours training, and DRs must accrue 100 CTCs across the three and a half years of their PhD.
The PhD candidate will have the unique opportunity to work collaboratively with the Loughborough AI research team in Computer Science for three years. They will have full access to specialist resources including laboratories, and High-Performance Computing facilities at Loughborough University and the expertise available within the Royal Botanic Gardens Kew team. The research will also provide opportunities to attend academic conferences, summer schools, and other training courses to improve technical skills. The candidate will master advanced deep learning techniques and have excellent career prospects on the successful completion of the PhD.
The student will be trained in techniques linked to data management and machine learning by supervisors Dr Haibin Cai. Training in trait based, sequencing and molecular systematics will be provided by Dr Eve Lucas, as well as laboratory and data technicians on site at Royal Botanic Gardens Kew.
Year 1: The candidate will conduct a comprehensive review of machine learning methods and large language models, with a focus on their application to trait extraction from herbarium specimens and scientific literature. They will meet regularly with supervisors to refine computer vision approaches and will develop algorithms for data pre-processing and image augmentation to improve recognition accuracy. The performance of these models will be evaluated across multiple herbarium datasets.
Year 2: The candidate is expected to submit a review paper to a peer-reviewed journal. They will consolidate a comprehensive fruit and seed trait database for Myrtaceae, combining manual data sources with AI-assisted extraction developed in Year 1. The first comparative phylogenetic analyses will be conducted, focusing on the evolution of fleshy fruits in relation to rainforest colonization and on trait-dependent diversification in Myrtaceae.
Year 3: The candidate will conduct large-scale comparative phylogenetic analyses across Myrteae and Syzygieae, including trait–environment correlations, models of geographic range size, and dispersal/colonization patterns in insular versus continental systems. They will also finalize the integration of AI pipelines with phylogenetic analyses, and focus on writing and submitting the main thesis chapters and manuscripts.
Triki, A., Bouaziz, B. and Mahdi, W., 2022. A deep learning-based approach for detecting plant organs from digitized herbarium specimen images. Ecological Informatics, 69, p.101590.
Goëau, H., Mora‐Fallas, A., Champ, J., Love, N.L.R., Mazer, S.J., Mata‐Montero, E., Joly, A. and Bonnet, P., 2020. A new fine‐grained method for automated visual analysis of herbarium specimens: A case study for phenological data extraction. Applications in Plant Sciences, 8(6), p.e11368.
Neotropical Myrtaceae Working Group, Staggemeier, V.G., Amorim, B., Bünger, M., Costa, I.R., de Faria, J.E.Q., Flickinger, J., Giaretta, A., Kubo, M.T., Lima, D.F. and dos Santos, L.L., 2024. Towards a species‐level phylogeny for Neotropical Myrtaceae: Notes on topology and resources for future studies. American Journal of Botany, 111(5), p.e16330.
Petrocelli, I., Alzate, A., Zizka, A. and Onstein, R.E., 2024. Dispersal‐related plant traits are associated with range size in the Atlantic Forest. Diversity and Distributions, 30(7), p.e13856.
Lu, L., Fritsch, P.W., Matzke, N.J., Wang, H., Kron, K.A., Li, D.Z. and Wiens, J.J., 2019. Why is fruit colour so variable? Phylogenetic analyses reveal relationships between fruit‐colour evolution, biogeography and diversification. Global Ecology and Biogeography, 28(7), pp.891-903.
For further information about this project, please contact Dr. Haibin Cai ([email protected]), www.lboro.ac.uk/departments/compsci/staff/haibin-cai/.
To apply to this project:
Applications must be submitted by 23:59 GMT on Wednesday 7th January 2026.