Global warming is causing crop insect pests to expand their geographic ranges and to undergo increased numbers of reproductive generations per year, with detrimental effects on agricultural crops. For instance, it is expected that an increase in global temperatures of 2°C will translate into a 46% increase in wheat yield losses due to additional insect pest damage.
Entomopathogenic (insect-infecting and killing) fungi represent a natural alternative to chemical pesticides as they can be exploited in sustainable biological control strategies, such as in the case of fungi of the genera Beauveria and Metarhizium. Entomopathogenic fungi live in ecologically rich and complex niches, including in terms of biodiversity, interactions, and evolutionary relationships, and have therefore developed the ability to produce a plethora of bioactive natural products, including insecticidal and immunosuppressive metabolites.
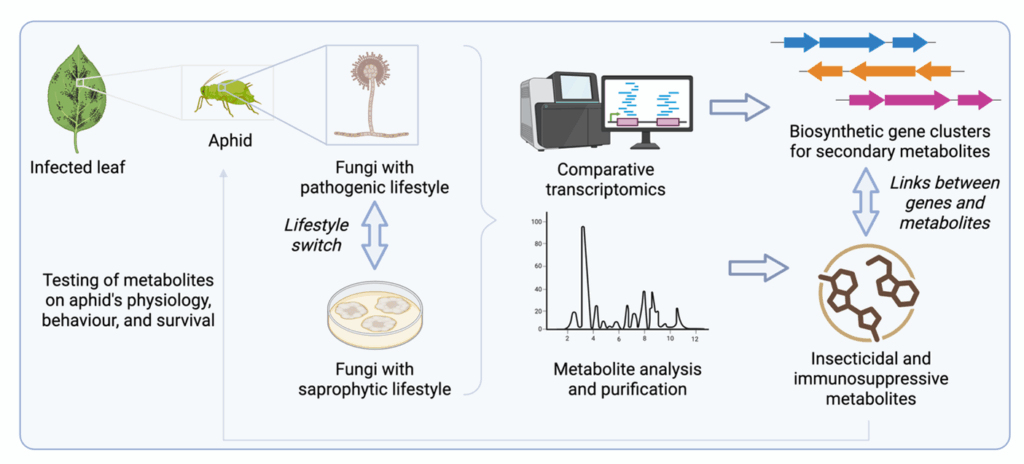
The ecological and biochemical interaction between fungi and herbivore insects remains understudied, particularly from the point of view of the genetic components that allow fungi to switch from a saprophytic to a pathogenic lifestyle. The aim of this PhD project is to characterise the ecological and biochemical interaction between entomopathogenic fungi and insects, using aphids as model organisms due to their short life cycles and their ecological relevance as widespread agricultural pests.
The focus of this project will be to investigate how fungi can perform the switch between saprophytic and pathogenic lifestyle. We anticipate that a variety of genes will be differentially expressed between the two lifestyles, including the upregulation of genes coding for proteases and for enzymes involved in secondary metabolite biosynthesis during infection. We will use this information to pinpoint biosynthetic gene clusters for secondary metabolites that enable entomopathogenic fungi to infect and kill insects, which will guide us in investigating the biosynthetic products made by these gene clusters and their effects on aphid physiology, behaviour, and survival. At the same time, we will investigate how infection from insects affects gene expression in aphids, where we anticipate observing cuticle response, proteasome and lysosome increased activity as well as upregulation of immune response pathways. Studying the aphid’s response to infection will provide further ecological insights into fungal-insect interactions.
This project is suitable for CASE funding
Each host has a slightly different application process.
Find out how to apply for this studentship.
All applications must include the CENTA application form.
Choose your application route
Comparative transcriptomic analyses: A selection of entomopathogenic fungi provided by the Fraunhofer Institute for Molecular Biology and Applied Ecology will be used to infect green peach aphids. Comparative RNAseq analysis will be performed on i/ fungi grown on culturing media, ii/ aphids infected by fungi, and iii/uninfected aphids, to identify genes that are differentially expressed in fungi that exhibit saprophytic vs. pathogenic lifestyle, as well as in fungal-infected vs. uninfected aphids.
Bioinformatic analyses: AntiSMASH and BlastP will be used to identify biosynthetic gene clusters putatively responsible for secondary metabolite biosynthesis, using the genomic sequences that we have already assembled through preliminary work.
Metabolite analyses: Flash chromatography and HPLC-guided fractionation of metabolite extracts from fungi will be used to purify secondary metabolites, followed by compound characterisation through mass spectrometry and NMR spectroscopy. Bioactivity testing of metabolites on aphids will allow us to study their effects on aphid physiology, behaviour, and survival.
DRs will be awarded CENTA Training Credits (CTCs) for participation in CENTA-provided and ‘free choice’ external training. One CTC can be earned per 3 hours training, and DRs must accrue 100 CTCs across the three and a half years of their PhD.
This is an interdisciplinary project at the interface between ecology, microbiology and chemistry. The student will receive specialist training in a variety of subjects including fungal biology, transcriptomics, bioinformatics, chromatography, mass spectrometry, and NMR spectroscopy.
The student will be supported to develop these skills by a team of supervisors with complementary expertise. They will also have access to the excellent facilities and support available in both the School of Life Sciences and the Department of Chemistry of the University of Warwick, e.g. high-performance computer clusters and Bioinformatics RTP for transcriptomics data analysis, UHPLC-HRMS and NMR spectroscopy instruments for metabolite analysis.
The Fraunhofer Institute for Molecular Biology and Applied Ecology (IME), a German publicly owned non-academic research institute, will be the CASE partner in this PhD project and will host the PhD student for a 6-month internship focused on fungal extracts metabolite identification and dereplication using a combination of metabolomics and bioinformatics analyses, including through specialised equipment available at their premises. Dr Marius Spohn, named PhD co-supervisor, is Deputy Head of the Department of Natural Products at the Fraunhofer IME.
Year 1: Fungal pathogenicity testing and transcriptomics (months 1-9). A selection of entomopathogenic fungal strains will be used to infect the green peach aphid, followed by comparative transcriptomics analyses, where RNA will be purified from i/ fungi grown on culturing media, ii/ aphids infected by fungi, and iii/uninfected aphids. This will allow us to identify genes that are differentially expressed in fungi that exhibit saprophytic vs. pathogenic lifestyle, as well as in fungal-infected vs. uninfected aphids.
Bioinformatic analyses (months 10-12). AntiSMASH and BlastP will be used to identify biosynthetic gene clusters putatively responsible for secondary metabolite biosynthesis, using genomic sequences that we have already assembled through preliminary work. RNA reads will be mapped against biosynthetic gene clusters to assess which ones are expressed during infection, and therefore likely to have insecticidal/immunosuppressive bioactivity.
Year 2: Metabolite analysis (months 13-24). Metabolites will be extracted from fungal strains grown in pure cultures and from aphids infected with the fungi, then analysed through UHPLC-HRMS. Molecular networking analysis (e.g. through GNPS) will be performed to aid metabolite identification and dereplication. Flash chromatography and HPLC-guided fractionation of metabolite extracts will be used to purify metabolites, followed by compound characterisation through mass spectrometry and NMR spectroscopy.
Year 3: Study of the ecological role of secondary metabolites (months 25-30). Bioactivity testing of purified metabolites will be performed on aphids to study their effects on aphid physiology, behaviour, and survival, providing insights into the ecological role of these molecules.
Thesis and manuscript write-up (months 31-36). The outcomes of the project will be written up in article style format, and manuscripts arising from the work will be submitted for publication to relevant journals.
Ma, C.-S., Wang, B.-X., Wang, X.-J., Lin, Q.-C., Zhang, W., Yang, X.-F., van Baaren, J., Bebber, D.P., Eigenbrode, S.D., Zalucki, M.P., Zeng, J., Ma, G., 2025. Crop pest responses to global changes in climate and land management. Nat Rev Earth Environ 6, 264–283. https://doi.org/10.1038/s43017-025-00652-3
Zhang, L., Fasoyin, O.E., Molnár, I., Xu, Y., 2020. Secondary metabolites from hypocrealean entomopathogenic fungi: Novel bioactive compounds. Nat Prod Rep 37, 1181–1206. https://doi.org/10.1039/c9np00065h
Jin, S., Alberti, F., 2025. Advances in the discovery and study of Trichoderma natural products for biological control applications. Nat Prod Rep 44, 367-1386. https://doi.org/10.1039/d5np00017c
Murindangabo, Y.T., Kopecký, M., Perná, K., Konvalina, P., Bohatá, A., Kavková, M., Nguyen, T.G., Hoang, T.N., 2024. Relevance of entomopathogenic fungi in soil–plant systems. Plant Soil 495, 287–310. https://doi.org/10.1007/s11104-023-06325-8
Reingold, V., Faigenboim, A., Matveev, S., Haviv, S., Belausov, E., Vilcinskas, A., Ment, D., 2024. Transcriptional reprogramming in the entomopathogenic fungus Metarhizium brunneum and its aphid host Myzus persicae during the switch between saprophytic and parasitic lifestyles. BMC Genomics 25, 917. https://doi.org/10.1186/s12864-024-10824-y
For any enquiries related to this project please contact Dr Fabrizio Alberti, [email protected].
To apply to this project:
Applications must be submitted by 23:59 GMT on Wednesday 7th January 2026.