Methane (CH4) accounts for ~30% of climate forcing. Trees are known to host methanotrophic bacteria yet the identity of the methane oxidising bacteria (MOB), their physiological characteristics and role of tree associated MOB driven methane oxidation for the global methane cycle are poorly understood at present. This applies to both bark and foliar MOB, with this project mainly focussing on the latter. CH4 uptake by trees is a known process, but the methanotrophs responsible remain poorly understood. Major challenges are the low abundance of methanotrophs, relatively low uptake rates and the difficulties encountered in their isolation. Foliar MOB may uptake atmospheric methane and thus potentially contribute to atmospheric methane removal but they are likely also a filter for soil and wood derived methane that is emitted via the leaves. The factors affecting tree MOB diversity and physiology are currently not established and the literature suggests that these are variable and may depend on a range of environmental factors. This project will work towards characterisation of the diversity, physiology and role of phyllosphere methanotrophs of diverse trees to gain a better understanding of their role in the global methane cycle.
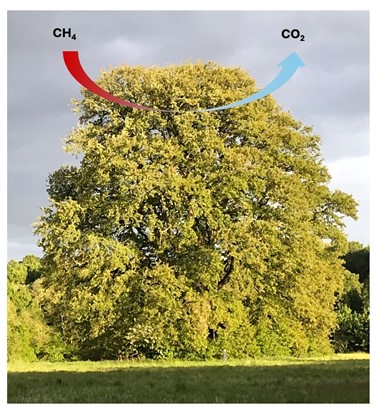
Figure 1: The role of methanotrophic (methane-oxidising) bacteria for the global methane cycle is poorly understood. One possibility is that high affinity atmospheric methane oxidising bacteria are taking up methane from the atmosphere at ~2ppmv concentration.
This project is not suitable for CASE funding
Each host has a slightly different application process.
Find out how to apply for this studentship.
All applications must include the CENTA application form.
Choose your application route
You will apply a combination of analytical, microbiological and molecular approaches in order to characterise the activity, diversity and physiology of tree associated methanotrophs. Gas chromatography will be applied to determine methane uptake rates of leaf samples or cells derived from leave biomass. Microbiological methodologies will include enriching and isolating methanotrophs from tree samples. Molecular techniques will include approaches for analysis of microbial diversity using metabarcoding as well as metagenomics or related techniques for cultivation-independent characterisation of tree MOB.
DRs will be awarded CENTA Training Credits (CTCs) for participation in CENTA-provided and ‘free choice’ external training. One CTC can be earned per 3 hours training, and DRs must accrue 100 CTCs across the three and a half years of their PhD.
Training in gas chromatography, handling gases and volatiles, microbiological and molecular approaches will be provided by the team of supervisors.
This project is a continuation of an established collaboration between researchers at Warwick (Schaefer, Bending) and Birmingham (Chen, Gauci, McDonald) with access to diverse expertise and the opportunity to access resources at both institutions including the Birmingham Institute for Forest Research (BiFoR).
Year 1: determination of methane uptake rates for diverse tree species over an annual cycle; establishment of enrichments and isolation procedures
Year 2: identification of MOB and investigation of their localisation on or within plant tissue; characterisation of MOB enrichments and potential isolates by (meta)genomic analysis
Year 3: Physiological characterisation of MOB; inoculation of tree samples with methanotroph cultures
GAUCI, V., PANGALA, S.R., SHENKIN, A. et al. 2024. Global atmospheric methane uptake by upland tree woody surfaces. Nature 631, 796–800. https://doi.org/10.1038/s41586-024-07592-w
GORGOLEWSKI, A. S., CASPERSEN, J. P., VANTELLINGEN, J. & THOMAS, S. C. 2023. Tree Foliage is a Methane Sink in Upland Temperate Forests. Ecosystems, 26, 174-186.
KEPPLER, F., HAMILTON, J. T. G., BRAS, M. & RÖCKMANN, T. 2006. Methane emissions from terrestrial plants under aerobic conditions. Nature, 439, 187-191.
PALMER, J. L., HILTON, S., PICOT, E., BENDING, G. D. & SCHÄFER, H. 2021. Tree phyllospheres are a habitat for diverse populations of CO-oxidizing bacteria. Environmental Microbiology, 23, 6309-6327.
Applicants are strongly encouraged to contact Hendrik Schäfer ([email protected]) to discuss the project and ask any questions they may have.
To apply to this project:
Applications must be submitted by 23:59 GMT on Wednesday 7th January 2026.