Project highlights
- Sewage pollution of rivers is also concerning because of the antimicrobial resistance (AMR) in bacteria carried with the sewage
- We want to understand the transport and fate of resistant bacteria in the environment
- In particular, we want to quantify the transmission of antimicrobial resistance via sewage into the environment
Overview
The widespread use of antibiotics in humans and animals has not only selected for bacteria carrying resistance genes but also for plasmids that can transfer these genes between different bacteria. This has led to a global spread of antimicrobial resistance (AMR), which has enormous public health implications for nations of all income and development levels. Analysing 88 pathogen-antibiotic combinations, 1·27 million (95% UI 0·911–1·71) deaths have been attributed to resistant infections globally in 2019. It is generally thought that exposure to untreated sewage is the major driver for the spread of resistance through the environment, but this has not been quantified and clinicians remain sceptical regarding the importance of the environment for AMR prevalence in the human population. Thus, it is important to have reliable, quantitative evidence to better inform risk analysis and policy guidance on this global health issue.
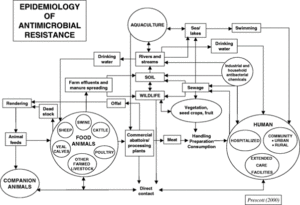
The input of untreated sewage to UK rivers has gained increased public and regulatory attention in the last few years and is a widespread problem largely owing to the dependence on very aged water infrastructure (https://theriverstrust.org/sewage-map). This is one pathway resistant bacteria can enter the environment, in addition to many others (Fig. 1) that include discharges from wastewater treatment plants, or as runoff from farms where animal manures and slurries are applied as fertilizers. Resistant bacteria may survive in river sediments that are then resuspended during flow events. Climate change is affecting this in at least two ways: by causing more frequent and severe flooding and also by causing droughts, which will in turn affect the transmission and fate of resistant bacteria.
This project aims to bring together disparate data to quantify the transmission of AMR via sewage into the environment, putting numbers on those arrows in Fig. 1 from/to sewage. This will involve data mining, statistical inference and mathematical modelling. Additional data can be measured to fill critical gaps in information or test hypotheses or predictions. This would enable the student to gain inter-disciplinary training and mentoring to tackle a critical link in the spread of AMR. The student would have a range of options to explore to provide the first quantitative assessment of how and when AMR can enter, persist, and be transported in river systems with different pollution sources.
Figure 1: One of the many schematics visualizing the network of AMR transmission in the environment. No scheme can be complete.
Host
University of BirminghamTheme
- Climate and Environmental Sustainability
- Organisms and Ecosystems
Supervisors
Project investigator
Jan-Ulrich Kreft (Biosciences, University of Birmingham, [email protected])
Co-investigators
Joshua Larsen (GEES, University of Birmingham, [email protected])
Co-I: Panayiota Touloupou (Maths, University of Birmingham, [email protected])
Co-I: Daniel Read (CEH, [email protected])
Including a wider supervisory team of collaborators in the AMRtransq network
How to apply
- Each host has a slightly different application process.
Find out how to apply for this studentship. - All applications must include the CENTA application form. Choose your application route
Methodology
A systematic review of existing data will be combined with developing methods that will allow us to combine measures of different entities (genes or bacteria) with different bases (presence/absence, frequency of positive samples, relative abundance, absolute abundance) to the extent possible. Based on these data, and any other data that will become available during the project, we will use Bayesian approaches and mathematical models to infer transmission rates where possible or use Bayesian Networks where only a combination of quantitative and more qualitative data is available. We will very likely find that some critical data is missing and will aim to measure these when possible. In addition, new measurements can be used to test predictions of our models. There is room to extend the scope of the work to include predicting effects of climate change or including ethnographic work to gather knowledge available from river water users and other citizens.
Training and skills
Students will be awarded CENTA2 Training Credits (CTCs) for participation in CENTA2-provided and ‘free choice’ external training. One CTC equates to 1⁄2 day session and students must accrue 100 CTCs across the three years of their PhD.
This project is highly interdisciplinary and will provide the student with a unique quantitative skills set including mathematical modelling, statistical data analysis, programming and potentially laboratory skills, enhanced by working with different partners.
This will be enhanced by collaboration with members of the AMR transdisciplinary networks that UKRI will fund and that will be open to new members. The networks will include all major stakeholders. Interactions with our collaborators on the AMRflows project in India offer further opportunities to gain insights and skills.
Moreover, the PhD student will have the opportunity for public outreach activities to inform the AMR debate with the results of our project. Project management and communication skills will also be gained.
Partners and collaboration
The research requires a wide range of expertise and access to data, so in addition to the core supervisory team based at the CENTA2 partners University of Birmingham and Centre for Ecology and Hydrology, we have a wider supervisory team of scientists who are part of the transdisciplinary network “AMRtransq: Quantifying transmission of AMR in natural and anthropogenic environments”.
Further details
Further details on how to contact the supervisor for this project and how to apply for this project can be found here:
For any enquiries related to this project please contact Jan-Ulrich Kreft ([email protected]).
To apply to this project:
- You must include a CENTA studentship application form, downloadable from: CENTA Studentship Application Form 2024.
- You must include a CV with the names of at least two referees (preferably three) who can comment on your academic abilities.
- Please submit your application and complete the host institution application process via: https://sits.bham.ac.uk/lpages/LES068.htm. Please select the PhD Bioscience (CENTA) 2024/25 Apply Now button. The CENTA application form 2024 and CV can be uploaded to the Application Information section of the online form. Please quote CENTA 2024-B27 when completing the application form.
Applications must be submitted by 23:59 GMT on Wednesday 10th January 2024.
Further information on supervisors and collaborators:
Dr Jan-Ulrich Kreft
School of Biosciences & Institute of Microbiology and Infection & Centre for Computational Biology
The University of Birmingham
Edgbaston, Birmingham, B15 2TT, UK
Tel: +44 (0)121 41-48851
Email: [email protected]
Web: https://more.bham.ac.uk/amrflows/ https://scholar.google.com/citations?user=hLRsYpsAAAAJ&hl=en&oi=ao
Dr Jan Kreft is a microbiologist and mathematical modeller with a wide range of experience in modelling the transfer or resistance plasmids, the population dynamics of sensitive and resistant bacteria in wastewater, wastewater treatment and rivers and using models combined with Bayesian inference to extract information from datasets.
Dr Joshua Larsen
School of Geography, Earth and Environmental Sciences
The University of Birmingham
Edgbaston, Birmingham, B15 2TT, UK
Email: [email protected]
Web: https://www.birmingham.ac.uk/staff/profiles/gees/larsen-joshua.aspx
Dr Josh Larsen is a hydrologist and has a wide range of expertise modelling transport of pollution in the environment and the use of remote sensing data.
Dr Panayiota Touloupou
School of Mathematics
The University of Birmingham
Edgbaston, Birmingham, B15 2TT, UK
Tel: +44 (0)1491 692644
Email: [email protected]
Web: https://www.birmingham.ac.uk/staff/profiles/maths/touloupou-panayiota.aspx
Dr Panayiota Touloupou is a statistician with a wide range of experience in mathematical modelling of infectious diseases and the development of novel Bayesian statistical methods needed for model fitting to data.
Dr Daniel Read
Centre for Ecology & Hydrology
Maclean Building, Benson Lane
Crowmarsh Gifford
Wallingford, Oxfordshire, OX10 8BB
Tel: +44 (0)1491 692644
Email: [email protected]
Web: https://www.ceh.ac.uk/staff/daniel-read
Dr Daniel Read has a number of large metagenomic datasets looking at wastewater and wastewater-river transitions that the student would be able to work on. Equally, Dr Read can support access to treatment works and river field sites in the Thames catchment where he has been working.
Professor Barbara Kasprzyk-Hordern
Department of Chemistry
Centre for Sustainable and Circular Technologies (CSCT)
Water Innovation and Research Centre (WIRC)
Centre for Therapeutic Innovation
Institute for Sustainability
Bath BA2 7AY, UK
Tel: +44 (0) 1225 385013
Email: [email protected]
Web: https://researchportal.bath.ac.uk/en/persons/barbara-kasprzyk-hordern
Professor Barbara Kasprzyk-Hordern <[email protected]> has been analysing pharmaceuticals such as antibiotics in wastewaters and developing methods to use prescription data to predict concentrations of pharmaceuticals in wastewater, which compare well with the measured values. She has extensively collaborated with water companies and contributed her expertise to wastewater based epidemiology
Professor Dov Stekel
Professor of Computational Biology
School of Biosciences
University of Nottingham
South Laboratory, Sutton Bonington Campus
Sutton Bonington, Leicestershire, LE12 5RD, UK
Tel: 0115 951 6294
Email: [email protected]
Web: https://www.nottingham.ac.uk/biosciences/people/dov.stekel
Professor Dov Stekel is a mathematician with extensive experience in modelling AMR dynamics and statistical inference and has developed models to quantify AMR transmission on farms.
Dr Lisa Avery
The James Hutton Institute
Craigiebuckler
Aberdeen AB15 8QH
Scotland UK
Tel: +44 (0)344 928 5428
Email: [email protected]
Web: https://www.hutton.ac.uk/staff/lisa-avery
Dr Lisa Avery leads the Centre for Human and Animal Pathogens in the Environment. Current projects include decentralised wastewater treatment and quantifying spatial AMR patterns in urban and rural landscapes.
Dr Rupert Hough
Head of Department Information and Computational Sciences
The James Hutton Institute (see above)
Email: [email protected]
Web: https://www.hutton.ac.uk/staff/rupert-hough
Dr Rupert Hough is a risk and exposure modeller specialising in systems approaches and brings specific expertise in quantifying relationships within human:ecological systems and ultimately the operationalisation of Figure 1 into a working model. Rupert also brings access to several national (Scotland) level datasets of resistance genes in both soils and waters that would help inform Figure 1, as well as access to site-specific data.
Dr Bing Guo
Senior Lecturer, Environmental Engineering
Fellow/Programme Lead at the Institute for Sustainability
School of Sustainability, Civil and Environmental Engineering
University of Surrey, Guildford GU2 7XH, UK
Tel: +44 (0)1483 686617
Email: [email protected]
Web: https://www.surrey.ac.uk/people/bing-guo
Dr Bing Guo is an environmental engineer working on anaerobic digestion, wastewater treatment, biofilms, microbial migration and AMR.
Dr Matthew Wade
Head of Science & Research
Environmental Monitoring for Health Protection
Analytics & Data Science Directorate
UK Health Security Agency
Email: [email protected]
Mobile: 07957708170
Web: www.gov.uk/ukhsa, https://researchportal.ukhsa.gov.uk/en/persons/matthew-wade
Dr Matt Wade has extensive experience of modelling Covid concentrations in wastewater and related aspects of wastewater based epidemiology.
Dr Isabel Douterelo
Lecturer Water and Applied Microbiology
Civil & Structural Engineering
The University of Sheffield
Sir Frederick Mappin Building , Sheffield, S1 3JD
Tel: +44 114 222 9728
Email: [email protected]
Web: https://www.sheffield.ac.uk/civil/people/academic/isabel-douterelo-soler
Dr Douterelo is an environmental scientist with broad experience in urban water pollution, her expertise lies in understanding how the microbiome associated with urban water infrastructure can affect public health and the environment. She can provide access to metagenomic data from sampling freshwater ecosystems (bathing site in Yorkshire) affected by faecal pollution, and access to urban water infrastructure research facilities at the University of Sheffield.
Dr Christopher Quince
Warwick Medical School – Microbiology and Infection
The University of Warwick, Coventry CV4 7AL, UK
Tel: +44 (0)2476 522317
Email: [email protected]
Web: https://warwick.ac.uk/fac/sci/med/staff/cquince/
High-Resolution Microbiomics Group Leader, Earlham Institute
Email: [email protected]
Web: https://www.earlham.ac.uk/profile/chris-quince
Dr Quince has extensive experience in mathematical modelling and statistical inference and his group has been at the forefront of developing more rigorous statistical and bioinformatic algorithms, e.g., for reconstructing genomes from metagenomes. Together with the Wellington lab at the University of Warwick, Dr Quince has driven forward the research on AMR dynamics in the Thames catchment with collaborators from CEH, Thames Water and others.
Possible timeline
Year 1
Systematic review of the existing evidence that could potentially be used to quantify transmission followed by development of a mathematical model and Bayesian methods to infer transmission parameters (Paper 1).
Having (very likely) identified gaps, we will decide which can be filled in by own work and the student will then make plans for the work to be included in years 2 and 3. Other gaps could be filled in by ongoing research of collaborators so part of the task is to communicate with collaborators on their plans and if they can help.
Year 2
This will likely be a mix of experimental work, collecting samples and analysing them in the lab, and bioinformatics of existing metagenomic datasets to infer changes in populations of resistant bacteria through the water cycle, for example examining whether resistance genes are lost from mobile genetic elements or not. To the extent that metagenomic reads can be assembled into MAGs (Metagenome Assembled Genomes), further analyses can be done. The bioinformatic analysis will lead to Paper 2.
Year 3
Work in year 3 will depend in a large part on progress made so far and progress achieved by collaborators in the transdisciplinary AMR networks that are being set up in 2024. It is expected that experimental work will continue with the aim to complete by the end of year 3 and that further data analysis will continue, leading to one paper on the experimental work and possibly one paper on the bioinformatics and modelling, or several smaller contributions to publications from collaborators.
Further reading
Hassoun-Kheir N, Stabholtz Y, Kreft JU, de la Cruz R, Romalde JL, Nesme J, Sørensen SJ, Smets BF, Graham D, Paul M (2020). Comparison of antibiotic-resistant bacteria and antibiotic resistance genes abundance in hospital and community wastewater: A systematic review. Science of The Total Environment 743: 140804
Arya S, Todman H, Baker M, Hooton S, Millard A, Kreft JU, Hobman JL, Stekel DJ (2020). A generalised model for generalised transduction: the importance of co-evolution and stochasticity in phage mediated antimicrobial resistance transfer. FEMS Microbiology Ecology 96: fiaa100
Amos GCA, Hawkey PM, Gaze WH, Wellington EM (2014). Waste water effluent contributes to the dissemination of CTX-M-15 in the natural environment. The Journal of Antimicrobial Chemotherapy 69: 1785–1791
Amos GCA, Zhang L, Hawkey PM, Gaze WH, Wellington EM (2014). Functional metagenomic analysis reveals rivers are a reservoir for diverse antibiotic resistance genes. Veterinary Microbiology 171: 441–447
Amos GCA, Gozzard E, Carter CE, Mead A, Bowes MJ, Hawkey PM, Zhang L, Singer AC, Gaze WH, Wellington EMH (2015). Validated predictive modelling of the environmental resistome. ISME Journal 9: 1467–1476
Lehmann K, Bell T, Bowes MJ, Amos GCA, Gaze WH, Wellington EMH, Singer AC (2016). Trace levels of sewage effluent are sufficient to increase class 1 integron prevalence in freshwater biofilms without changing the core community. Water Research 106: 163–170
Singer AC, Shaw H, Rhodes V, Hart A (2016). Review of Antimicrobial Resistance in the Environment and Its Relevance to Environmental Regulators. Frontiers in Microbiology 7: 1728
Quintela-Baluja M, Frigon D, Abouelnaga M, Jobling K, Romalde JL, Gomez Lopez M, Graham DW (2021). Dynamics of integron structures across a wastewater network – Implications to resistance gene transfer. Water Research 206: 117720
Lee K, Raguideau S, Sirén K, Asnicar F, Cumbo F, Hildebrand F, Segata N, Cha C-J, Quince C (2023). Population-level impacts of antibiotic usage on the human gut microbiome. Nature Communications 14: 1191