Project highlights
- Multidisciplinary science – This project combines biodiversity science, bioinformatics, and artificial intelligence to develop cost-effective tools for the real-time monitoring of biodiversity by end-users.
- Big data science – This project uses advanced computational tools and biostatistics to identify impact of industrial processes (e.g. wastewater treatment) on freshwater ecosystems. These actions enable regulatory compliance and sustain healthy ecosystems.
- Link to industry – The co-design and co-supervision of the PhD by Severn Trent Water ensures that wet lab and computational tools developed by the DR are suitable for real-world applications by end-users. These tools enable regulatory compliance by industry and will enable real-time assessment of production processes on biodiversity.
Overview
Biodiversity is declining at an alarming rate, affecting the delivery of ecosystems services on which we all rely for our well-being and the economy. These services include climate regulation, food provisioning, clean water and recreation, to name a few. With the exit from the EU, the UK established the 25-year environment plan to improve environmental quality within a generation, and the Environment Act 2021 to maintain biodiversity net gain while enabling growth. These necessary regulations demand a rapid response from industry to assess the impact of production processes on biodiversity to meet compliance. In this climate, cost-effective and rapid biodiversity monitoring is critical for effective biodiversity conservation, resource management, and compliance.
Traditional biodiversity monitoring techniques, such as direct observations require specialist taxonomic skills, are not standardised when it comes to sampling protocols, can be invasive, are low throughput and biased towards species that leave identifiable remains. Conversely, DNA-based methods (e.g. metabarcoding) have revolutionised conventional biodiversity research by enabling high throughput screening from environmental matrices without being limited to species with well-preserved remains. These methods are based on the detection of species through the analysis of the DNA they shed in environmental matrices (environmental DNA). However, DNA-based techniques require outsourcing to high throughput sequencing facilities with turnaround time of up to eight weeks, delaying decision-making related to industrial processes.
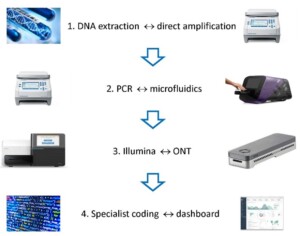
In this project the DR will develop a portable toolbox for onsite biodiversity assessment to overcome current limitations in biodiversity monitoring. This will be achieved by deploying a field-based toolkit for the real-time monitoring of biodiversity within 24h. The field-based laboratory will use microfluidics and the portable Oxford Nanopore Sequencing Technology (ONT) for real-time assessment of biodiversity, including in remote and protected areas. The mobile laboratory will be validated in a case study that assesses the impact of wastewater effluent on river biodiversity in the UK. In addition, the DR will develop a user-friendly analytical dashboard for end-users allowing the real-time assessment of the impact of production processes on biodiversity. The team combines the knowledge base expertise in developing novel tools for biodiversity monitoring and analytical tools with the end-user insights on the biodiversity toolbox, accelerating implementation of science-driven solutions.
Figure 1: Key steps to enable field-based real-time biodiversity monitoring by end-users.
CENTA Flagship
This is a CENTA Flagship Project
Case funding
This project is suitable for CASE funding
Host
University of BirminghamTheme
- Organisms and Ecosystems
Supervisors
Project investigator
Prof Luisa Orsini, University of Birmingham and The Alan Turing Institute, [email protected]
Co-investigators
Pu Xia, School of Biosciences, University of Birmingham ([email protected])
Jiarui Zhou, School of Biosciences, University of Birmingham ([email protected])
Cynthia Carliell-Marquet, Severn Trent Water ([email protected])
How to apply
- Each host has a slightly different application process.
Find out how to apply for this studentship. - All applications must include the CENTA application form. Choose your application route
Methodology
Wetlab skills. The DR will learn the application of high-throughput sequencing technologies to environmental DNA extracted from environmental matrices.
Biocomputing. Metabarcoding or marker gene sequencing will be collected from rivers downstream of wastewater plants. The DR will learn to analyse these data using bioinformatics and multivariate statistics.
- AI. Sparse Canonical Correlation Analysis (sCCA) will be employed to regress measures of biodiversity attributes on biotic and abiotic factors in standard, multiple regressions using generalized linear models (GLM). sCCA is ideal for discovering complex, group-wise patterns between high-dimensional datasets.
Translation. To create long-lasting impacts beyond the project, the DR will have placements with the water industry where they will engage with the innovation and commercial teams to ensure transfer of knowledge and drive translation of research findings into end-user applications.
Training and skills
Students will be awarded CENTA2 Training Credits (CTCs) for participation in CENTA2-provided and ‘free choice’ external training. One CTC equates to 1⁄2 day session and students must accrue 100 CTCs across the three years of their PhD.
The DR will receive multidisciplinary training spanning from biodiversity science (Orsini), computational biology (Xia), and AI (Zhou). The DR will benefit from access to some of the most up to date high performance computing facilities in the country, including UoB and Alan Turing facilities. The DR will spend up to 6 months at Severn Trent Water to complete a case study and learn the skills of translational science.
Partners and collaboration
This is a CASE application in which the Severn Trent offers funding at £1K per year, and placements for the DR in the applied innovation team. Dr Carliell-Marquet will co-supervise the DR. She is an architect at Severn Trent in the Innovation team who’s remit is Wastewater Systems, covering all aspects of wastewater collection, treatment and disposal including the impact of wastewater on the ecological health of waterbodies. Dr Carliell-Marquet is the author of the ‘Enhancing Nature’ Innovation Strategy for Severn Trent Water, within which she is driving the introduction of next generation biodiversity monitoring and the development of Ecological Digital Twins for river catchments at Severn Trent. She has co-designed the proposed project and will offer co-supervision and training to the DR.
Further details
Further details on how to contact the supervisor for this project and how to apply for this project can be found here:
For any enquiries related to this project please contact Prof Luisa Orsini, [email protected].
To apply to this project:
- You must include a CENTA studentship application form, downloadable from: CENTA Studentship Application Form 2024.
- You must include a CV with the names of at least two referees (preferably three) who can comment on your academic abilities.
- Please submit your application and complete the host institution application process via: https://sits.bham.ac.uk/lpages/LES068.htm. Please select the PhD Bioscience (CENTA) 2024/25 Apply Now button. The CENTA application form 2024 and CV can be uploaded to the Application Information section of the online form. Please quote CENTA 2024-B37 when completing the application form.
Applications must be submitted by 23:59 GMT on Wednesday 10th January 2024.
Possible timeline
Year 1
Year 1: development of a field-based toolkit for real time-assessment of biodiversity using eDNA metarbarcoding and oxford nanopore technology. Student conference in Birmingham. Write literature review. Draft publication on technological developments.
Year 2
Year 2: collection of samples from UK rivers downstream of wastewater plants agreed with the water industry for a case study. Application of bioinformatics and multivariate statistics to analyse the data. Application of AI tools to identify drivers of biodiversity loss. Draft empirical paper.
Year 3
Year 3: Placement with Severn Trent. Develop and test the user-friendly dashboard for end-user application. Write thesis. Attend international conference.
Further reading
- Eastwood N, Stubbings WA, Abdallah MA, Durance I, Paavola J, Dallimer M, Pantel JH., Johnson S, Zhou J, Hosking JS, Brown JB, Ullah S, Krause S, Hannah DM, Crawford SE, Widmann M, Orsini L (2022) The Time Machine framework: monitoring and prediction of biodiversity loss’. Trends in Ecology and Evolution (Invited opinion paper) 37: 138-146. (https://doi.org/10.1016/j.tree.2021.09.008)
- Eastwood N., Zhou J., Derelle R., Jia Y., Crawford S., Abdallah M., Davidson T. A., Brown J. B., Hollert H., Colbourne J.K., Creer S., Bik H., Orsini L. (2023) 100 years of anthropogenic impact causes changes in freshwater functional biodiversity. eLife (in press) (https://doi.org/10.1101/2023.02.26.530075)
- Nogués-Bravo D, Rodríguez-Sánchez F, Orsini L, de Boer E, Jansson R,Morlon H, Fordham DA, Jackson ST (2018) Cracking the Code of Biodiversity Responses to Past Climate Change Trends in Ecology and Evolution (Invited review) 33: 765-776 (https://doi.org/10.1016/j.tree.2018.07.005)
- Bohmann, K., Evans, A., Gilbert, M. T., Carvalho, G. R., Creer, S., Knapp, M., . . . de Bruyn, M. (2014). Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol Evol, 29(6), 358-367. doi:10.1016/j.tree.2014.04.003