Project highlights
- This project will identify genes that might control oak ageing, and test if they are the same genes or not that control ageing in short-lived plants.
- This project will attempt to produce ‘fast-cycling’ oak for speed-breeding purposes.
- This project will test the effect of future climates on living oaks, in collaboration with BIFoR.
Overview
Can oaks survive climate change? Oak (Quercus robur) is a significant UK tree species, providing both timber and animal feed crops (acorns), and in ecological terms it is a keystone species, with each tree a habitat for up to 200 other species (Mitchell et al., 2019). Oak has evolved to be a remarkably long-lived and slow-growing tree compared to its close relatives, with young trees taking 40-100 years to begin flowering (Vieitez et al., 2012). This means that today’s generation of oak trees will bear the brunt of the changing climate, without giving oak populations time to adapt to the environmental and disease-based challenges this will present. Evidence is emerging that other tree species are responding to climate change by altering the time at which they flower (Büntgen et al., 2022), risking the potential loss of entire species if plant-pollinator interactions are disrupted or trees cannot otherwise reproduce. The potential risk of climate change to oak must therefore be taken seriously, but large-scale experiments such as the Free Air Carbon Enrichment (FACE) study being conducted by the Birmingham Institute for Forest Research (BIFoR) allow us to simulate future atmospheric conditions and predict the challenges facing oak in the future.
We do not yet understand why oak takes so long to reproduce, because plant ageing mechanisms have been studied only in fast-growing annual plants such as the fast-growing laboratory plant model Arabidopsis thaliana (Wu et al., 2009). If oak ageing could be artificially accelerated then humanity will have a powerful new tool to help oak survive the coming climate crisis. This project proposes to take the first steps towards this goal in the following ways:
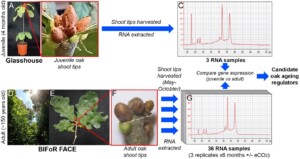
- Using existing unpublished oak RNA-seq datasets (Figure 1), conduct bioinformatic analyses to identify the genes that control ageing and flowering oak, and test if these are similar to the plant ageing mechanisms known from Arabidopsis through wet-lab experiments.
- Use oak genetic engineering technologies (Álvarez & Ordás 2007) to try to speed up ageing in oak saplings.
- Determine how future atmospheric CO2 concentrations will affect the fitness of today’s oaks, utilising the FACE experiment at BIFoR.
Figure 1: Candidate oak ageing regulators will be identified by comparing existing unpublished RNA-seq data. Shoot tips were harvested from oak saplings (A,B). Messenger RNA (mRNA) was extracted from these and sequenced (C). Shoot tips were harvested from adult trees at BIFoR FACE (D-F) across the growing season, and mRNA extracted and sequenced (G). Genes with a consistent difference in expression between juvenile and adult samples are candidate regulators of ‘adulthood’, ie. competency to flower.
Host
University of BirminghamTheme
- Organisms and Ecosystems
Supervisors
Project investigator
Andrew Plackett, University of Birmingham, a.r.g.p[email protected]
Co-investigators
Graeme Kettles, University of Birmingham, [email protected]
Richard Buggs, Royal Botanic Gardens, Kew, [email protected]
How to apply
- Each host has a slightly different application process.
Find out how to apply for this studentship. - All applications must include the CENTA application form. Choose your application route
Methodology
This project provides the opportunity to learn and use a wide range of techniques:
The student will use bioinformatics to analyse existing data from an oak RNA-seq experiment to identify genes that could control oak ageing. They will test these by checking their expression by quantitative real-time PCR (qPCR) across a 3-year library of samples already available in the lab.
To test if the same ageing mechanisms exist in oak as in the model plant Arabidopsis thaliana, the student will genetically engineer Arabidopsis to insert oak versions of known ageing genes and see if they have the same function.
The student will help to set up an oak transformation system based on previously-published tissue culture techniques and somatic embryogenesis to create genetically engineered oak.
There is the opportunity to undertake live field-sampling at the BIFoR FACE experiment or to undertake phenotypic analysis of archive material previously collected from the experiment.
Training and skills
Students will be awarded CENTA2 Training Credits (CTCs) for participation in CENTA2-provided and ‘free choice’ external training. One CTC equates to 1⁄2 day session and students must accrue 100 CTCs across the three years of their PhD.
Training in all project-specific techniques can be provided by the project supervisors. Prof Richard Buggs can provide support with bioinformatic analyses. Dr Graeme Kettles can provide training and support and with qPCR. Dr Andrew Plackett can provide training and support in tissue culture, the genetic engineering of Arabidopsis and field-sampling at the FACE experiment. Other transferrable-skills training is available via the University of Birmingham.
It would be advantageous for the student to have some prior experience with bioinformatic analysis and general wet-lab experience with molecular biology techniques. A valid driver’s licence accepted within the UK would also be an advantage.
Partners and collaboration
Richard Buggs is a Professor of Evolutionary Genomics at the Queen Mary University London, and leads the leads the Plant Health and Adaptation research team at Royal Botanic Gardens Kew. His research group analyses genome sequences to understand how plants, especially trees, adapt in response to climate change and new pests and pathogens. It is envisaged that this project will be based primarily at the University of Birmingham Edgbaston campus with Prof Buggs providing advice and support remotely.
Further details
Further details on how to contact the supervisor for this project and how to apply for this project can be found here:
For any enquiries related to this project please contact Andrew Plackett, a.r.g.p[email protected].
To apply to this project:
- You must include a CENTA studentship application form, downloadable from: CENTA Studentship Application Form 2024.
- You must include a CV with the names of at least two referees (preferably three) who can comment on your academic abilities.
- Please submit your application and complete the host institution application process via: https://sits.bham.ac.uk/lpages/LES068.htm. Please select the PhD Bioscience (CENTA) 2024/25 Apply Now button. The CENTA application form 2024 and CV can be uploaded to the Application Information section of the online form. Please quote CENTA 2024-B38 when completing the application form.
Applications must be submitted by 23:59 GMT on Wednesday 10th January 2024.
Possible timeline
Year 1
Undertake bioinformatic analysis to identify oak candidate ageing genes and testing by qPCR. Identify and clone oak versions of Arabidopsis genes for use in genetic engineering. Set up and test oak tissue-culture protocol.
Year 2
Create genetically-engineered Arabidopsis plants carrying oak versions of ageing genes and establish pure-breeding lines for analysis. Attempt genetic engineering of oak. Conduct live FACE field sampling or collect data from archive material. Glasshouse experiments testing CO2 enrichment on oak saplings.
Year 3
Test genetically-engineered Arabidopsis lines for oak gene functions in controlling ageing. Test genetically-engineered oak saplings for accelerated ageing by qPCR or RNA-seq. Complete analysis of FACE field/archive samples.
Further reading
Journals:
Álvarez, R. and Ordás, J. J. (2007) ‘Improved genetic transformation protocol for cork oak (Quercus suber L.)’, Plant Cell, Tissue and Organ Culture, 91, pp. 45-52. doi: 10.1007/s11240-007-9276-6.
Büntgen, U., Piermattei, A., Krusic, P. J., Esper, J., Sparks, T., and Crivellaro, A. (2022) “Plants in the UK flower a month earlier under recent warming”, Proceedings of the Royal Society B., 289, article 20212456. doi: 10.1098/rspb.2021.24.
Mitchell, R. J., Bellamy, P. E., Ellis, C. J., Hewison, R. L., Hodgetts, N. G., Iason, G. R., Littlewood, N. A., Newey, S., Stockan, J. A., and Taylor, A. F. S. (2019) ‘Collapsing foundations: The ecology of the British oak, implications of its decline and mitigation options’, Biological conservation, 233, pp. 316-327. doi: 10.1016/j.biocon.2019.03.040
Vieitez, A. M., Corredoira, E., Martínez, M. T., San-José, M. C., Sánchez, C., Valledares, S., Vidal, N., and Ballester, A. (2012) ‘Application of biotechnological tools to Quercus improvement’, European Journal of Forest Research, 131, pp. 519-539. doi: 10.1007/s10342-011-0526-0.
Wu, G., Park, M. W., Conway, S. R., Wang, J. W., Weigel, D., and Poethig, R. S. (2009) ‘The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis’, Cell, 138, pp. 750-759. DOI: 10.1016/j.cell.2009.06.031.
Web-page:
BIFoR FACE experiment. Available at:
https://www.birmingham.ac.uk/research/bifor/face/index.aspx (Accessed: 4 September 2023)