Project highlights
- Multidisciplinary science – This project combines life and computer science. Within computer science, it applies cutting-edge AI technologies such as digital twins, deep learning, graph machine learning, and temporal data modelling. Within the life sciences, it applies high throughput sequencing technologies, bioinformatics and biostatistics.
- Big data science – This project uses advanced computational methodologies and biostatistics to predict biodiversity loss under different climate and pollution scenarios. It can be applied to assess the potential impact of industrial processes on biodiversity.
- Link to industry – This PhD was co-designed by an applicant from Nature Matrics, a world-leading eDNA company, with a focus on the translation of fundamental science into the commercial applications of tools and approaches developed by the DR. These tools can also be applied by regulators to manage biodiversity loss.
Overview
Biodiversity is declining at an alarming rate, affecting the delivery of ecosystem services on which we all rely for our well-being and the economy. These services include climate regulation, food provisioning, clean water and recreation, to name a few. With the exit from the EU, the UK established the 25-year environment plan to improve environmental quality within a generation, and the Environment Act 2021 to maintain biodiversity net gain while enabling growth. These necessary regulations demand a rapid response from industry to assess the impact of production processes on biodiversity. Yet, we lack the tools to predict the impact of industrial processes on natural biological diversity, thereby limiting our ability to conserve critical resources and the services they provide. One of the main challenges that conservationists face is predicting how severe biodiversity loss is and what are the main drivers of this loss.
Traditional methods for species dynamics – species distribution models, population viability analyses, and statistical learning techniques – focus on individual species, missing by design species interactions within the food web and with the environment. Holistic, ecosystem-level approaches are largely missing because they are challenging to develop and apply.
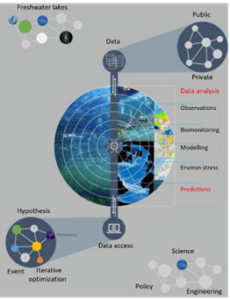
In this project, the DR will develop state-of-the-art tools to monitor and predict biodiversity loss under different climate and pollution scenarios. The DR will apply Temporal Graph Networks (TGNs) to temporal biodiversity data collected from freshwater ecosystems using environmental DNA. The TGN is a cutting-edge AI model and will be used to create a digital twin model for freshwater biodiversity. The digital twin will integrate historical records of eDNA and environmental data to dynamically model their interactions over time. It will offer multi-scale, holistic modelling that tracks changes across taxonomic groups over time and space and links them to environmental change, identifying the main drivers of loss. The digital twin will use these temporal trends to predict biodiversity loss under business as usual and restoration plans. The digital twin for biodiversity has the potential to become the tool of choice for biodiversity management and conservation.
To make the tool accessible to end-users, the DR will develop an intuitive analytical dashboard. This dashboard will allow the direct assessment of production processes, land use and other human activities on biodiversity. By synergistically combining advanced computational and bioinformatics technologies with end-user insights, the team aims to accelerate the transition from traditional to science-driven, technologically enhanced solutions for biodiversity conservation.
Figure 1: The framework that uses digital twins to monitor and predict biodiversity loss under different climate and pollution scenarios. The lake biodiversity Digital Twin uses AI-supported data fusion, data analytics and predictions to improve biodiversity management.
Host
University of BirminghamTheme
- Organisms and Ecosystems
Supervisors
Project investigator
Dr Jiarui (Albert) Zhou, School of Biosciences, ([email protected])
Co-investigators
Prof Luisa Orsini, University of Birmingham and The Alan Turing Institute, ([email protected])
Dr Shan He, University of Birmingham, ([email protected])
How to apply
- Each host has a slightly different application process.
Find out how to apply for this studentship. - All applications must include the CENTA application form. Choose your application route
Methodology
Biocomputing. Metabarcoding or marker gene sequencing data will be collected from freshwater ecosystems to capture community-level biodiversity. The DR will learn to analyse these data using bioinformatics and multivariate statistics.
AI. Temporal Graph Networks (TGNs) will be employed to model dynamic changes in biodiversity over time and space in response to environmental factors. Designed to model graph-structured data that evolve over time, TGNs can effectively capture temporal dependencies and node interactions, facilitating monitoring and prediction of biodiversity loss.
Digital Twins. Digital twins will be constructed as a data-driven representation of real-world ecosystems. The biodiversity digital twin will integrate environmental factors and biodiversity quantified with eDNA, utilising TGNs to dynamically model their interactions, offering multi-scale and multi-taxa predictions.
Translation. To create long-lasting impacts beyond the project, the DR will design a user-friendly interface that will be tested and applied to real-world cases by potential industry partners like Nature Metrics to ensure the transfer of knowledge and drive the translation of research findings into end-user applications.
Training and skills
Students will be awarded CENTA2 Training Credits (CTCs) for participation in CENTA2-provided and ‘free choice’ external training. One CTC equates to 1⁄2 day session and students must accrue 100 CTCs across the three years of their PhD.
The DR will receive multidisciplinary training spanning from AI and modelling (Zhou and Shan) and biodiversity science (Orsini). The student will join the research activities in the cross-college Centre for Environmental Research and Justice (CERJ) at the UoB, benefiting from the training, collaborations, and resources it provides in support of environmental science research. The DR will benefit from access to some of the most up-to-date high-performance computing facilities in the country, including UoB and Alan Turing facilities. They will learn the skills of translational science.
Partners and collaboration
This PhD synergies with our case study in the Horizon Europe PARC project which focuses on biodiversity monitoring and prediction. The student will collaborate with our PARC partners in the UK and EU on eDNA data processing and computational modelling. The PhD student may also work closely with our collaborators in industry and government agencies, including Nature Matrics (eDNA), Environment Agency (environmental data), and UK Centre for Ecology & Hydrology (molecular ecology).
Further details
Further details on how to contact the supervisor for this project and how to apply for this project can be found here:
For any enquiries related to this project please contact Dr Albert Zhou ([email protected])
To apply to this project:
- You must include a CENTA studentship application form, downloadable from: CENTA Studentship Application Form 2024.
- You must include a CV with the names of at least two referees (preferably three) who can comment on your academic abilities.
- Please submit your application and complete the host institution application process via: https://sits.bham.ac.uk/lpages/LES068.htm. Please select the PhD Bioscience (CENTA) 2024/25 Apply Now button. The CENTA application form 2024 and CV can be uploaded to the Application Information section of the online form. Please quote CENTA 2024-B48 when completing the application form.
Applications must be submitted by 23:59 GMT on Wednesday 10th January 2024.
Possible timeline
Year 1
Bioinformatics analysis of high throughput sequencing data generated using eDNA metabarcoding. These data are extant at the Co-I Orsini’s laboratory. Student conference in Birmingham. Write the literature review.
Year 2
Application of Temporal Graph Networks (TGNs) and other AI models to identify drivers of biodiversity loss under current and future scenarios of pollution and climate change. Draft empirical paper.
Year 3
Working with potential industry partners. Develop and test the user-friendly dashboard for the end-user application. Write thesis. Attend international conferences.
Further reading
- Eastwood N, Stubbings WA, Abdallah MA, Durance I, Paavola J, Dallimer M, Pantel JH., Johnson S, Zhou J, Hosking JS, Brown JB, Ullah S, Krause S, Hannah DM, Crawford SE, Widmann M, Orsini L (2022) The Time Machine framework: monitoring and prediction of biodiversity loss’. Trends in Ecology and Evolution (Invited opinion paper) 37: 138-146. (https://doi.org/10.1016/j.tree.2021.09.008)
- Eastwood N., Zhou , Derelle R., Jia Y., Crawford S., Abdallah M., Davidson T. A., Brown J. B., Hollert H., Colbourne J.K., Creer S., Bik H., Orsini L. (2023) 100 years of anthropogenic impact causes changes in freshwater functional biodiversity. eLife (in press) (https://doi.org/10.1101/2023.02.26.530075)
- Rossi E., Chamberlain B., Frasca F., Eynard D., Monti F., & Bronstein M. (2020). Temporal graph networks for deep learning on dynamic graphs. arXiv preprint arXiv:2006.10637. (https://doi.org/10.48550/arXiv.2006.10637)
- Borowiec M. L., Dikow R. B., Frandsen, P. B. McKeeken, A. Valentini, G. & White A. E. (2022). Deep learning as a tool for ecology and evolution. Methods in Ecology and Evolution, 13(8), 1640-1660. (https://doi.org/10.1111/2041-210X.13901)
- Jeong K. S., Kim D. K., Jung J. M., Kim M. C., Joo G. J. (2008). Non-linear autoregressive modelling by Temporal Recurrent Neural Networks for the prediction of freshwater phytoplankton dynamics. ecological modelling, 211(3-4), 292-300 (https://doi.org/10.1016/j.ecolmodel.2007.09.029)