Project highlights
- We will test if trees have a molecular memory of past environmental stresses
- We will apply most advanced DNA sequencing approaches to investigate woods
- For the first time, we will explore relations between epigenetic and dendrochronology data in trees
Overview
Trees are long living, sessile organisms that represent a fundamental component of most of earth ecosystems. Being immobile during their life, trees need to efficiently adapt to changes of environment and to stresses, in order to reach maturity and reproduction. However, contrary to animals, trees do not rely on a learning strategy based on a central-nervous apparatus, and the existence of certain form of adaptive memory in these organisms is debated.
Recent advanced in genomics studies are always more convincingly showing that environmental information can be recorded at molecular level on DNA through epigenetic marks. Epigenetics studies molecular changes of DNA (mostly DNA methylation) in response to developmental or environmental stimuli, which does not involve a change of the DNA sequence itself. These changes can be inherited during cell division and are particular stable in plants, where they represent the base of an epigenetic “memory”, which remains encoded in biological tissues and can be transmitted to progenies.
In trees, wood is produced by plant cells that lignified and die to provide physical functions such as of mechanical support, storage and water conduction. However, part of their DNA is preserved unaltered, and retains the epigenetic information encoded in the cells in the moment they died. In temperate zones, most trees produce in their wood one growth-ring each year, so that the entire period of a tree’s life remain impressed in its wood material. As wood growth is affected by environmental conditions, trees have marks of past interactions with the environment recorded into their wood as they grow, at both physical and molecular level. However, while physical propriety of tree rings is well investigated, the possible epigenetic changes associated to the residual DNA in different rings re.
This project aims to understand how trees use environmental information to adapt to a changing condition. This will be done by extracting DNA from different parts of tree rings and compare their epigenetic profiles. The PhD researcher will compare the DNA methylation from different tree rings and associate epigenetic changes to historical and ecological record of drought. This will allow to connect tree molecular and physical responses to the environment in in tree rings, creating for the first time a direct connection among dendrochronology, ecology and epigenetic data in trees.
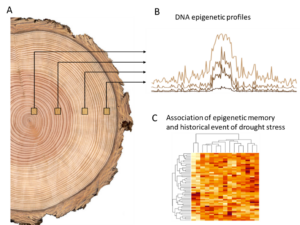
Figure 1: graphical representation of the project design. A) DNA is extracted from wood sampled at different part of a trees ring. B) the samples are sequenced and a whole genome DNA methylation profile is generated and compared. C) differences in DNA methylation are associated to historical event of drought stress.
Host
University of BirminghamTheme
- Organisms and Ecosystems
Supervisors
Project investigator
Marco Catoni (UoB [email protected])
Co-investigators
Adriane Esquivel-Muelbert (UoB – [email protected])
How to apply
- Each host has a slightly different application process.
Find out how to apply for this studentship. - All applications must include the CENTA application form. Choose your application route
Methodology
The project will require to merge molecular, computational, and ecological methodologies. We will adapt newly developed protocols of DNA extraction from oak wood to produce libraries for high-throughput sequencing analysis, to generate genome-wide single cytosine resolution maps of DNA methylation of wood cells located in different portion of a tree ring sample.
Then, we will analyse tree-rings from oak samples located at the BIFoR facility (University of Birmingham), for which historical records of past drought stress are available. We will determined physical changes in wood associated to drought stress and we will determine the associated portion of epigenetic changes found in each ring portion.
Finally, we will compare and validate our result with analysis performed on oak seedling drought stressed in controlled condition (greenhouses), to determine a final set of DNA regions that can be used to track past event of water deprivation in oak wood.
Training and skills
Students will be awarded CENTA2 Training Credits (CTCs) for participation in CENTA2-provided and ‘free choice’ external training. One CTC equates to 1⁄2 day session and students must accrue 100 CTCs across the three years of their PhD.
The candidate would join the School of Biosciences and the School of Geography, Earth and Environmental Sciences where we have a vibrant community of researchers working on diverse plant species. Specifically, the student will be directly supervised by two PIs, and will benefit from peer-discussion occurring during weekly lab meeting sessions. We also have close connections with the Centre for Computation Biology, which provides further expertise in genomic analysis. Additionally, the student will have access to training programmes run by the University, these include training in bioinformatics and career development workshops for postgraduate students organised through our graduate school.
Further details
Further details on how to contact the supervisor for this project and how to apply for this project can be found here:
For any enquiries related to this project please contact Marco Catoni, [email protected].
To apply to this project:
- You must include a CENTA studentship application form, downloadable from: CENTA Studentship Application Form 2024.
- You must include a CV with the names of at least two referees (preferably three) who can comment on your academic abilities.
- Please submit your application and complete the host institution application process via: https://sits.bham.ac.uk/lpages/LES068.htm. Please select the PhD Bioscience (CENTA) 2024/25 Apply Now button. The CENTA application form 2024 and CV can be uploaded to the Application Information section of the online form. Please quote CENTA 2024-B6 when completing the application form.
Applications must be submitted by 23:59 GMT on Wednesday 10th January 2024.
Possible timeline
Year 1
Based on a previous developed protocol of DNA extraction from wood, designed for high throughput sequencing, you will develop a procedure for BS-seq (bisulfite DNA treatment and sequencing) starting from oak tree rings. The student will produce and compare single cytosine methylation profiles from multiple replicates and we will optimise the computational pipeline to determine differentially methylated regions in Oak genome.
Year 2
You will collect different tree rings from oaks from the BIFoR site, and collect and analysed ecological data of the area. The method established in year 1 will be then used to produce the DNA methylation profiles of different tree rings (ideally, a sequenced library collected at 4-5 regions per sample). Then you will use different epigenetic analysis (clustering, PCA) to determine the variability across replicates and significant epigenetic differences which are specific of certain tree ring areas.
Year 3
You will integrate precipitation over the last 50 years at the BIFoR site with the variation in the epigenetic profile observed in wood area formed at different years, and will determine a core of chromosomal areas that can store molecular memory of drought stress in trees.
Further reading
Zeng, Z., Raffaello, T., Liu, M.-X., and Asiegbu, F.O. (2018). Co-extraction of genomic DNA & total RNA from recalcitrant woody tissues for next-generation sequencing studies. Future Science OA 4, FSO309.