Project highlights
- Unravelling the intricate interactions of microbes in natural, complex environments.
- Understanding the origins of antimicrobial resistance (AMR).
- Tracing AMR transport via microbial genomes across ecosystems.
Overview
Globalization, growing and increasingly mobile human populations, intensive farming, chemical pollution, ecosystem degradation, and climate change have led to the emergence of new microbial pathogens and the spread of antimicrobial resistance (AMR). Considering this, there are concerns that connectivity between host and environmental microbiome reservoirs may play a role in the ongoing AMR pandemic, contributing to more than a million deaths per year globally. The origins of AMR, however, lay in the complex interactions of microbial communities in natural environments. An important aspect of AMR occurrence is its transport between biomes and hosts, which is often ignored. AMR transport not only contributes to processes influencing microbial community assembly but is relevant in light of the One Health concept designed to counteract the global health challenges associated with the movement and evolution of AMR.
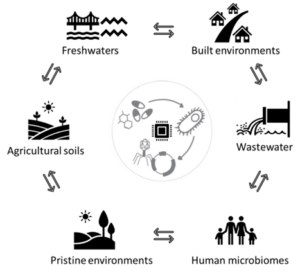
Figure 1: Tracking of antimicrobial resistance (AMR) transport, mediated via bacterial genomes and mobile genetic elements across the land-water interface, including natural and built environments.
For this PhD studentship, you will characterise and track AMR genes across different natural and built environments. These include, but are not limited to, assessing AMR in the microbial fraction of soil (natural/pristine and agricultural), freshwater (rivers and lakes), wastewater environments and publicly available host-associated gut microbiomes. The land-water interface, although hypothesised as playing an important role, has rarely been assessed in the context of AMR transport, this adding novelty to the proposed research. This project will employ molecular methods including metagenomic DNA sequencing, coupled with onsite/field sampling, wet- and dry-lab techniques. The goals of this research are three-fold as outlined below:
- Characterise patterns of AMR occurrence and diversity within pristine and anthropogenically influenced soils and freshwaters.
- Generate a metagenomically-derived genome catalogue to explore the links between microbial taxa (MAGs) across biomes.
- Track the transport of AMR genes via MAGs at the land-water interface and vice versa.
CENTA Flagship
This is a CENTA Flagship Project
Host
UK Centre for Ecology & HydrologyTheme
- Organisms and Ecosystems
Supervisors
Project investigator
Dr. Susheel Bhanu Busi, UKCEH
Co-investigators
Dr. Holly Tipper, UKCEH
Dr. Gary D. Bending, University of Warwick
How to apply
- Each host has a slightly different application process.
Find out how to apply for this studentship. - All applications must include the CENTA application form. Choose your application route
Methodology
The project will comprise of a mix of fieldwork, wet- and dry-lab methodologies. Data from national scale, cross-biome surveys such as the Environment Agency’s River Surveillance Network and UKCEH’s Countryside Survey will be used for this project. There will also be the opportunity to collect and analyse microbiomes from across wastewater, freshwater and soil interfaces. DNA extraction will be done using established protocols and metagenomic sequencing libraries will be generated, including long-read DNA sequencing to characterise microbiomes. Dry-lab methodologies including the processing of metagenomic data from reads to the generation of MAGs will be done via existing and validated pipelines. The AMR profiles of the MAGs will be obtained using tools designed to leverage on the updated Comprehensive Antibiotic Resistance Database (CARD). Population genetics methodologies including strain tracking based on single nucleotide polymorphisms will be used to understand the transport of AMR along the land-water and vice versa interface.
Training and skills
Students will be awarded CENTA2 Training Credits (CTCs) for participation in CENTA2-provided and ‘free choice’ external training. One CTC equates to 1⁄2 day session and students must accrue 100 CTCs across the three years of their PhD.
The student will receive comprehensive training, including experimental design and sample selection, molecular wet-lab techniques (DNA extraction, PCR, sequencing library prep), and dry-lab methods (sequence data processing, assembly, MAG generation). Emphasis will be on functional annotation, especially identifying AMR genes and their taxonomic context. Skills encompass sample handling, bioinformatics, data visualization, statistics, interdisciplinary collaboration, ethical research, and literature review. The research will equip them for metagenomic research, ensuring proficiency in both laboratory and computational aspects, fostering responsible and reproducible research practices, and facilitating collaboration in diverse scientific fields.
Partners and collaboration
The student will benefit from a close association with the project CASE partner (the Environment Agency, TBC) and their ongoing research into developing new approaches for assessing ecosystem health. The Environment Agency are in the process of producing benchmark datasets to examine microbial communities across multiple taxonomic groups from 1000s of biofilm samples collected from rivers across England. The student will have access to these data, as well as to stored extracted DNA, allowing re-analysis and comparisons with alternative DNA sequencing approaches, including long-read technologies.
Further details
Further details on how to contact the supervisor for this project and how to apply for this project can be found here:
For any enquiries related to this project please contact Dr Susheel Bhanu Busi, [email protected].
The successful applicant would be registered at the University of Warwick.
To apply to this project:
- You must include a CENTA studentship application form, downloadable from: CENTA Studentship Application Form 2024.
- You must include a CV with the names of at least two referees (preferably three) who can comment on your academic abilities.
- Please submit your application and complete the host institution application process via: https://warwick.ac.uk/fac/sci/lifesci/study/pgr/studentships/nerccenta/ Complete the online application form – selecting course code P-C1PB (Life Sciences PhD); from here you will be taken through to another screen where you can select your desired project. Please enter “NERC studentship” in the Finance section and add Nikki Glover, [email protected] as the scholarship contact. Please also complete the CENTA application form 2024 and submit via email to [email protected]. Please quote CENTA 2024-UKCEH2 when completing the application form.
Applications must be submitted by 23:59 GMT on Wednesday 10th January 2024.
Possible timeline
Year 1
Over three years, the PhD will focus on accomplishing academic and research goals, including literature review and lab skills development in Year 1. In Year 2, it will emphasize laboratory work, data analysis, and presentation of findings. Year 3 involves advanced data analysis, thesis preparation, and paper submissions. Additionally, personal and professional development activities will be undertaken from years 1-3, including networking, joining societies, teaching, leadership roles, and workshops on job searches, grant writing, project management, and self-care. These activities will foster holistic growth, ensuring the candidate’s readiness for a successful post-PhD career.
Year 2
See above.
Year 3
See above.
Further reading
- Cornforth, D. M. and K. R. Foster (2015). “Antibiotics and the art of bacterial war.” Proceedings of the National Academy of Sciences 112(35): 10827-10828.
- Liguori, K., et al. (2022). “Antimicrobial Resistance Monitoring of Water Environments: A Framework for Standardized Methods and Quality Control.” Environmental Science & Technology 56(13): 9149-9160.
- Niehus, R., et al. (2021). “The evolution of strategy in bacterial warfare via the regulation of bacteriocins and antibiotics.” eLife 10: e69756.