Project highlights
- Tropical Tree Systematics
- Mobilisation of taxonomic data for species identification
- Machine Learning
Overview
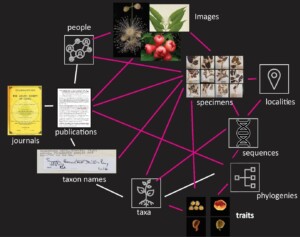
Aggregated vouchered plant occurrence data and associated images available in user facing portals enable easier access and reuse but many data and literature resources necessary for accurate identification remain scattered. Initiatives like the digital extended specimen (Fig. 1 adapted from Page 2013) that integrate heterogenous data (including distribution, phylogenetics and micromorphology) across institutional boundaries are starting to work out how to address this data disconnect but are still in the planning stages.
Taxonomic output provides immensely valuable training data through e.g., the updating of specimen identifications, classifying specimens at different taxonomic ranks and enumerating their biotic and abiotic characteristics and variables (traits). This project produce such data through revision of a section of the world’s largest tree genus, Syzygium (Myrtaceae), through classical taxonomy, molecular systematics and study of range and functional traits. Syzygium (Low et al. 2022) is a highly appropriate model group for such a study as it is considerably species rich (~1,200 accepted names), but hugely understudied and in need of thorough revisionary work. The greatest gap in Syzygium systematics is in Syzygium subgenus Syzygium (c. 1000 species), this subgenus has recently been divided into informal ‘sections’ including the lineage including the type of the genus, Syzygium sect. Syzygium (est. 500 species). Recent study has proposed diagnostic characters for that section.
This project aims to develop an extensible digital working environment, delivering image-based, robust, and taxonomically authoritative identification of plants at a global scale for biodiversity monitoring purposes (e.g., plot-based inventory and conservation assessment), to mobilise and interact with data at scale, and to recognise new species. Identifications enabled in the tool will be strengthened by explicit reference to currently scattered metadata associated with herbarium specimen images. Much of this associated information is currently linked only in the human brain; this tool will make these relationships explicitly available to other stakeholders. Integrating different kinds of data in this way makes it more explicit. Surfacing implicit data is a key aim of open science and will be an asset for training multi-modal machine learning models. Machine learning is a fast-moving field, the tool will enable integration with new approaches as they become available. This project will investigate effective ways to fuse the textual and visual information by combining large language models (LLMs) and Convolutional Neural Networks (CNNs) for clustering Syzygium species.
Figure 1: Existing (white lines) and future links (red) between different data classes for the tree genus Syzygium. Data classes linked in this way have been termed the ‘digital extended specimen’ .
CENTA Flagship
This is a CENTA Flagship Project
Host
UK Centre for Ecology & HydrologyTheme
- Climate and Environmental Sustainability
- Organisms and Ecosystems
Supervisors
Project investigator
Dr Oliver Pescott, UK Centre for Ecology & Hydrology. [email protected]
Co-investigators
Dr Haibin Cai, University of Loughborough. [email protected]
Dr Eve Lucas, Royal Botanic Gardens Kew. [email protected]
Dr Nicky Nicolson, Royal Botanic Gardens Kew. [email protected]
How to apply
- Each host has a slightly different application process.
Find out how to apply for this studentship. - All applications must include the CENTA application form. Choose your application route
Methodology
Determining which taxa are included in Syzygium sect. Syzygium will be achieved by systematic survey of all nomenclatural types of species in this section (c. 1000), first using existing machine learning algorithms for clustering followed by manual checking of resulting clusters. This hypothesis will be tested by building a molecular phylogenetic reconstruction of the appropriate cluster (c. 500 sequences) using high throughput baited sequencing. Trait and other data classes (e.g. geography, phylogeny, habitat, habit, use) will be accessed and linked using Echinopscis, a that enables easy collate digital information from separate resources using open science practices. Resulting Syzygium data, identification keys and automatic identification tools will be mobilised online via an appropriate digital portal such as WFO or POWO, as well as in a traditional taxonomic format. The checklist will serve as a ‘living monograph’ of a group that currently represents a major part of the taxonomic impediment.
Training and skills
Students will be awarded CENTA2 Training Credits (CTCs) for participation in CENTA2-provided and ‘free choice’ external training. One CTC equates to 1⁄2 day session and students must accrue 100 CTCs across the three years of their PhD.
The student will have the opportunity for training via staff and resources at the University of Loughborough, the Centre for Ecology & Hydrology and the Royal Botanic Gardens Kew. Skills obtained will be in the fields of i. data mobilisation and management for systematics and classification, ii. machine learning for automatic species identification and iii. herbarium taxonomy.
Partners and collaboration
The student will be trained in techniques linked to data management and machine learning by supervisors Dr Oliver Pescott, Dr Nicky Nicolson and Dr Haibin Cai. Training in trait based, sequencing and molecular systematics will be provided by Dr Eve Lucas, as well as laboratory and data technicians on site at Royal Botanic Gardens Kew.
Further details
Further details on how to contact the supervisor for this project and how to apply for this project can be found here:
For any enquiries related to this project please contact Dr Oliver Pescott, UK Centre for Ecology & Hydrology. [email protected], or Dr Eve Lucas, Royal Botanic Gardens Kew. [email protected].
The successful applicant would be registered at Loughborough University.
To apply to this project:
- You must include a CENTA studentship application form, downloadable from: CENTA Studentship Application Form 2024.
- You must include a CV with the names of at least two referees (preferably three) who can comment on your academic abilities.
- Please submit your application and complete the host institution application process via: https://www.lboro.ac.uk/study/postgraduate/apply/research-applications/ The CENTA application form 2024 and CV can be uploaded at Section 10 “Supporting Documents” of the online portal. Under Section 4 “Programme Selection” the proposed study centre is Central England NERC Training Alliance. Please quote CENTA 2024-UKCEH3-LU11 when completing the application form.
- For further enquiries about the application process, please contact the School of Social Sciences & Humanities ([email protected]).
Applications must be submitted by 23:59 GMT on Wednesday 10th January 2024.
Possible timeline
Year 1
The first year of the project will focus on determining which taxa are included in Syzygium sect. Syzygium. This will be achieved by systematic survey of all nomenclatural types of species in this section (c. 1000), first using existing machine learning algorithms for clustering followed by manual checking of resulting clusters. This hypothesis will be tested by building a molecular phylogenetic reconstruction of the final cluster (c. 500 sequences) using high throughput baited sequencing.
Year 2
Year two of the project explores assembling trait data and other data classes (e.g. geography, phylogeny, habitat, habit, use) to test and develop auto-identification using machine learning. Imaged herbarium specimens will be assembled from Kew, Leiden, Singapore, FRIM and other herbaria that hold sizable Southeast Asian Syzygium collections, as well as the global image aggregator GBIF.
Year 3
Year three of the project will utilize the outputs of the phylogenetic systematics, data digitally linked from the Auto-ID tool using Echinopscis to compile a comprehensive checklist of Syzygium sect. Syzygium that will be mobilised online via an appropriate digital portal such as WFO or POWO, as well as in a traditional taxonomic format. The checklist will serve as a ‘living monograph’ of a group that currently represents a major part of the taxonomic impediment.
Further reading
Journal:
Low, Y.W., Rajaraman, S., Tomlin, C.M., Ahmad, J.A., Ardi, W.H., Armstrong, K., Athen, P., Berhaman, A., Bone, R.E., Cheek, M. and Cho, N.R., (2022). Genomic insights into rapid speciation within the world’s largest tree genus Syzygium. Nature communications, 13(1), p.5031.
Nicolson, N., & Lucas, E. (2023). The Role of the OLS Program in the Development of echinopscis (an Extensible Notebook for Open Science on Specimens). Biodiversity Information Science and Standards, 7, 53.
Page, R.D., (2013). BioNames: linking taxonomy, texts, and trees. PeerJ, 1, p.190.
Web page with an author:
Nicolson, D. (2023) Echinopsis An extensible notebook for open science on specimens https://echinopscis.github.io/