Project highlights
- Reconstruction of ancient maize genomes from archaeological samples
- Apply state of the art plant genomics techniques
- Join a large project team investigating maize evolution in South America
Overview
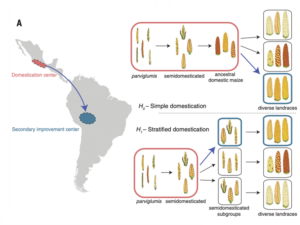
Maize was domesticated around 9,000 years ago in the Balsas region of Mesoamerica, whereupon it spread to both South and North America. Recent ground-breaking research in this group established that this occurred in several complex waves (Kistler et al 2018 Science 362:1309–1313). Through the use of ancient DNA to generate genomes from archaeological maize samples it was established that maize most likely left the domestication centre in an incompletely domesticated state and that landraces were subsequently formed outside the domestication centre in a ‘stratified’ domestication, Figure 1. This finding predicts the existence of semi-wild forms of maize being used in early agriculture far away from the area of origin. The existence of such samples has yet to be confirmed, and it is unknown how long such types may have persisted. In recent years archaeological samples of apparently semi-domesticated maize have been unearthed in Brazil, dating to around 1000 years ago. In this project we will recover archaeogenomes from these maize samples, determine genetically their domestication status and establish whether these samples support the stratified domestication hypothesis. This project will join a larger NERC funded team investigating maize in South America with a post-doctoral worker, research assistant and a global team of experts in archaeogenomics and archaeology in South America.
Figure 1: The stratified domestication model of maize .
Host
University of WarwickTheme
- Organisms and Ecosystems
Supervisors
Project investigator
Professor Robin G Allaby, University of Warwick
Co-investigators
Dr Logan Kistler (Smithsonian Institute, USA)
Dr Fabio Freitas (Embrapa, Brazil)
How to apply
- Each host has a slightly different application process.
Find out how to apply for this studentship. - All applications must include the CENTA application form. Choose your application route
Methodology
Archaeogenomes will be obtained from archaeological maize samples by retrieving ancient DNA using a dedicated ancient DNA laboratory in the University of Warwick, and subject to NGS sequencing techniques such as NovaSeq. Genomes will then be subject to bioinformatic and population genetics analyses. The allelic state of over 200 known domestication associated loci will be assessed to determine the level of domestication in samples. The genomes of these samples will be placed in the wider population structure of South America using a range of population genomic techniques.
Training and skills
Students will be awarded CENTA2 Training Credits (CTCs) for participation in CENTA2-provided and ‘free choice’ external training. One CTC equates to 1⁄2 day session and students must accrue 100 CTCs across the three years of their PhD.
The student will be trained in ancient DNA techniques such as extraction, library preparation (single stranded) and analysis of next generation sequencing (NGS) data. The student will gain state of the art genomics skills. Training in coding skills will also be provided in high level languages such as Perl, Python and R. The student will also partake in workshops on the wider NERC project.
Partners and collaboration
The student will receive co-supervision from Dr Kistler at the Smithsonian Institute. This will include at least one short visit to the Smithsonian and group there to gain skills.
Further details
Further details on how to contact the supervisor for this project and how to apply for this project can be found here:
For any enquiries related to this project please contact Professor Robin G Allaby, [email protected].
To apply to this project:
- You must include a CENTA studentship application form, downloadable from: CENTA Studentship Application Form 2024.
- You must include a CV with the names of at least two referees (preferably three) who can comment on your academic abilities.
- Please submit your application and complete the host institution application process via: https://warwick.ac.uk/fac/sci/lifesci/study/pgr/studentships/nerccenta/ Complete the online application form – selecting course code P-C1PB (Life Sciences PhD); from here you will be taken through to another screen where you can select your desired project. Please enter “NERC studentship” in the Finance section and add Nikki Glover, [email protected] as the scholarship contact. Please also complete the CENTA application form 2024 and submit via email to [email protected]. Please quote CENTA 2024-W16 when completing the application form.
Applications must be submitted by 23:59 GMT on Wednesday 10th January 2024.
Possible timeline
Year 1
Initial ancient DNA retrieval and screening of samples. Selection of samples to take forward to genome sequencing, initial generation of archaeogenomes by NovaSeq.
Year 2
Bioinformatic processing of archaeogenomes, mapping to modern genomes. Population structure based analyses, analysis of domesticate gene content.
Year 3
Commence thesis writing. Continued genome analysis, preparation of papers. Presentation of findings at international conferences (e.g. ISBA).
Further reading
Allaby RG, Stevens CJ, Kistler L, Fuller DQ (2021) Emerging evidence of plant domestication as a landscale-level process. Trends Ecol. Evol. 37:268-279.
Kistler K, Maezumi SY, Gregorio de Souza J, Przelomska NAS, Costa FM, SmithO, Loiselle H, Ramos-Madrigal J, Wales N, Ribeiro ER, Morrison RR, Grimaldo C, Prous AP, Arriaza B, Gilbert MTP, Oliveira Freitas F, Allaby RG (2018) Multiproxy evidence highlights a complex evolutionary legacy of maize in South America Science 362:1309–1313
Kistler L, Thaker HB, VanDewarker AM, Domic A, Berström A, George RJ, Harper TK, Allaby RG, Hirth K, Kennett DJ (2020) Archaelogical Central American maize genomes suggest ancient gene flow from South America. Proc. Natl. Acad. Sci. U.S.A. 117:33124-33129
Smith O, Nicholson WV, Kistler L, Mace E, Clapham A, Rose P, Stevens C, Ware R, Samavedam S, Barker G, Jordan D, Fuller DQ, Allaby RG (2019) A domestication history of dynamic adaptation and genomic deterioration in Sorghum. Nature Plants 5:369-379.
COVID-19
If another period of lockdown should occur, much of the work of this project could be carried out remotely after initial DNA processing. This part of the work could be carried out in a relatively short time period (ie 2 months). Reduction in contact would be achieved by cancelling international trips.