Project highlights
- developing methods and infrastructure for in-field sequencing of eDNA by citizen scientists
- improving community ability to assess biodiversity
- enabling monitoring of species that might otherwise be intractable, e.g. cryptic, nocturnal, rare
Overview
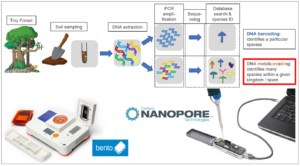
Accurately surveying biodiversity is of key importance as natural environments come under increasing threat. DNA-based methods now allow biodiversity assessment via environmental DNA (eDNA) analysis of soils, sediments, seas, freshwater, and air. Typically, eDNA is extracted from biological materials, ‘barcode’ segments are PCR amplified, these are sequenced using Illumina high-throughput technologies, and then compared to reference databases of barcode sequences. This ‘metabarcoding’ approach provides a catalogue of genera or species present, allowing assessment of species living in an environment, following changes in species composition through time, and monitoring of invasive or rare species. For some species, the only realistic way to identify and track their presence is by searching for their DNA.
Citizen science also plays an important role in upscaling biodiversity monitoring globally through programs such as eBird, City Nature Challenge, iNaturalist, Plant Alert. To date, citizen scientists have been involved in eDNA sampling and metadata collection (e.g. CALeDNA). Challenges for eDNA citizen science programs include maintaining sample quality between collection and receiving it at the lab, backlogs in sequencing samples due to time and funding pressures, and maintaining citizen engagement. Technological advances could now allow citizen scientists to process eDNA samples in the field, submit data to a platform and deliver rapid results to citizen scientists, solving many of the challenges.
Earthwatch is a global expert in citizen science and environmental monitoring. In the UK, Earthwatch’s citizen science programs centre on research in freshwater, urban and agricultural environments and have engaged thousands of people in monitoring the health and functioning of their local environments. This project aims to use Earthwatch’s citizen science networks and Oxford Nanopore technology to develop methods by which citizen scientists can sequence the eDNA samples they collect and receive results rapidly, facilitating environmental monitoring, reporting, and action.
Figure 1: Schematic illustration of environmental sampling, DNA extraction and PCR approaches to species identification.
Host
University of LeicesterTheme
- Organisms and Ecosystems
Supervisors
Project investigator
- Dr Celia A May (University of Leicester, [email protected])
Co-investigators
Dr Claire Narraway (Earthwatch, [email protected])
Prof Mark A Jobling (University of Leicester, [email protected])
Dr Moya L Burns (University of Leicester, [email protected])
How to apply
- Each host has a slightly different application process.
Find out how to apply for this studentship. - All applications must include the CENTA application form. Choose your application route
Methodology
- Collection of soil and freshwater samples from a range of environments, in collaboration with Earthwatch and their citizen scientists;
- Extraction of eDNA using appropriate commercial kits;
- Agnostic analysis of eDNA in samples via metabarcoding using portable nanopore sequencing;
- Targeted analysis of particular species via directed barcoding based on local ecosystem knowledge;
- Bioinformatic processing of barcode and metabarcode sequence data;
- Liaising with citizen scientists to explore barriers facing implementation of eDNA approaches; development of surveys/questionnaires and analysis of data;
- Liaising with Earthwatch colleagues to consider development of user-friendly analysis platforms to promote dissemination of technologies.
Training and skills
DRs will be awarded CENTA Training Credits (CTCs) for participation in CENTA-provided and ‘free choice’ external training. One CTC can be earned per 3 hours training, and DRs must accrue 100 CTCs across the three and a half years of their PhD.
Specific training: DNA extraction; PCR; gel electrophoresis; nanopore sequencing; bioinformatic data processing and interpretation; collaborator and citizen scientist liaison; formulation, implementation, analysis of surveys/questionnaires.
The DR will be embedded within the University of Leicester research culture and engage in discussions and presentations throughout their study. They will be encouraged to apply for independent field-work grants, e.g. via the Genetics Society, to help support testing with citizen scientists.
The candidate will work with Earthwatch, gaining skills in engaged environmental research and network widely through participation in the international Miyawaki Research Network, exploring the benefits and challenges of the Miyawaki afforestation method.
Partners and collaboration
Earthwatch “connects people with scientists worldwide to conduct environmental research and empowers them with the knowledge they need to conserve the planet.” This project will be co-supervised by Dr Claire Narraway, Senior Researcher Earthwatch Europe, and who is directly involved in the Tiny Forests project. She will facilitate interactions with citizen scientists and colleagues to a) provide suitable sources of eDNA, b) examine barriers to implementation of in-field nanopore sequencing & c) development of a user-friendly analysis platform. She will complement the UoL supervisory team consisting of geneticists with experience of nanopore sequencing (CAM, MAJ), and an ecologist (MLB).
Further details
For informal enquiries, please contact Dr Celia A May ([email protected]).
To apply to this project:
- You must include a CENTA studentship application form, downloadable from: CENTA Studentship Application Form 2025.
- You must include a CV with the names of at least two referees (preferably three) who can comment on your academic abilities.
- Please submit your application and complete the host institution application process via: CENTA PhD Studentships | Postgraduate research | University of Leicester. Please scroll to the bottom of the page and click on the “Apply Now” button. The “How to apply” tab at the bottom of the page gives instructions on how to submit your completed CENTA Studentship Application Form 2025, your CV and your other supporting documents to your University of Leicester application. Please quote CENTA 2025-L11 when completing the application form.
Applications must be submitted by 23:59 GMT on Wednesday 8th January 2025.
Possible timeline
Year 1
Development of a species-specific barcoding assay (e.g. for tracing greater crested newt) that can be performed with the portable lab equipment and nanopore sequenced using a flongle. Field trial with Earthwatch citizen scientists.
Year 2
Comparison of nanopore sequencing metabarcoding performance with “gold standard” Illumina sequencing. Development of survey/questionnaire to understand barriers to implementation of eDNA sequencing in the field by citizen scientists.
Year 3
Deployment and interpretation of survey completed by Earthwatch citizen scientists. Consideration of requirements and possible development needs for a user-friendly analysis platform in conjunction with Earthwatch colleagues. Thesis writing.
Further reading
- Earthwatch: https://earthwatch.org.uk
- Donald J, et al.: Multi-taxa environmental DNA inventories reveal distinct taxonomic and functional diversity in urban tropical forest fragments. Global Ecol Conserv 2021, 29:e01724. doi.org/10.1016/j.gecco.2021.e01724
- Thomsen PF, Willerslev E: Environmental DNA – An emerging tool in conservation for monitoring past and present biodiversity. Biol Conservation 2015, 183:4-18. doi.org/10.1016/j.biocon.2014.11.019
- Nanopore sequencing: https://nanoporetech.com/platform/technology