Project highlights
- Identify anaerobic bacteria degrading a pivotal atmospheric trace gas
- Use the identity of candidate DMS degraders to select environmental genomes for bioinformatic analysis
- Characterise anaerobic DMS degradation pathways in isolated strains using -omics approaches
Overview
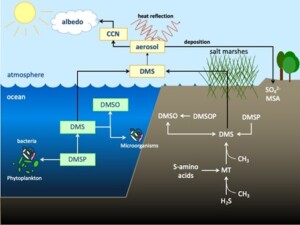
Dimethylsulfide (DMS) is a volatile sulfur compound of wide environmental significance. It has been described as the ‘smell of the sea’, due to the large emissions of DMS from marine environments into the atmosphere, but DMS sources also exist in terrestrial and freshwater environments. It is an important precursor for atmospheric aerosols which have major influence on radiative forcing and sulfur transport between the marine and terrestrial environments. DMS production and degradation are intimately linked to the activities of microorganisms. Various microbial processes are responsible for production of DMS from different precursor compounds and metabolic processes (Schäfer et al., 2010). Major sources of DMS include the degradation of dimethylsulfonium propionate (DMSP), the methylation of methanethiol (MT), respiratory reduction of dimethylsulfoxide (DMSO), which itself is produced by degradation of dimethylsulfoxonium propionate (DMSOP). Microorganisms utilise DMS as a carbon, sulfur and/or energy source in oxic and anoxic environments. Previously isolated bacteria (from anoxic marine sediments and thermophilic fermenters) shown to degrade DMS anaerobically coupled dissimilation of DMS to reduction of sulfate and nitrate (Tanimoto and Bak, 1994, Visscher and Taylor, 1993, Lyimo et al., 2009), but the enzymatic and genetic basis of DMS degradation in these or any anaerobic bacteria has not been reported. Incubation experiments with peatland samples have demonstrated anaerobic degradation of DMS in incubations with added ferric iron, in addition to sulfate and nitrate (Haaijer et al., 2008). These previous studies suggest that anaerobic degradation of DMS is due to diverse microorganisms and a major sink for DMS, but currently the diversity of these anaerobic microorganisms and their underlying metabolic diversity is poorly understood.
Figure 1: Environmental role and cycling of dimethylsulfide (DMS) and related compounds, dimethylsulfonium propionate (DMSP), dimethylsulfoxonium propionate (DMSOP), dimethylsulfoxide (DMSO), methanethiol (MT). The emission of DMS to the atmosphere provides aerosol precursors and allows transport of oxidation products like sulfate (SO42-) and methanesulfonate (MSA) to terrestrial environments (modified from (Kappler and Schäfer, 2014)).
Host
University of WarwickTheme
- Organisms and Ecosystems
Supervisors
Project investigator
- Prof Hendrik Schaefer (University of Warwick), [email protected]
Co-investigators
- Dr Ozge Eyice (University of Birmingham)
How to apply
- Each host has a slightly different application process.
Find out how to apply for this studentship. - All applications must include the CENTA application form. Choose your application route
Methodology
The microorganisms driving anaerobic DMS degradation will be characterised using cultivation-dependent and -independent approaches. Samples of anoxic soils and sediments will be amended with a range of different electron acceptors and DMS degradation will be monitored by gas chromatography. Microbial diversity of incubations will be assessed by 16S rRNA amplicon sequencing, selected samples will be used for 13C-DMS stable isotope probing and meta-omic analyses. Candidate DMS degrading microorganisms will be further investigated based on metagenome assembled genomes (MAGs) and metagenomic and metatranscriptomic datasets including those from previous work of the supervisory team (Tebbe et al., 2023, Tsola et al., 2024). Isolation of DMS-degrading organisms will also be pursued. Isolates will be characterised, genome sequenced, and specific enzymes and genes induced on DMS will be identified by proteomic and/or transcriptomic analyses.
Training and skills
DRs will be awarded CENTA Training Credits (CTCs) for participation in CENTA-provided and ‘free choice’ external training. One CTC can be earned per 3 hours training, and DRs must accrue 100 CTCs across the three and a half years of their PhD.
Training in the labs of Schäfer and Eyice will provide the student with opportunities to learn new methodologies or deepen their current experience and understanding of a wide range of microbiological and environmental microbiology methods (meta-omics), bioinformatics and chemical analytical methods including environmental sampling, stable isotope probing, gas and ion chromatography, extraction of DNA, RNA, proteins, 16S rRNA amplicon sequencing, transcriptomics, proteomics.
Partners and collaboration
This project will be a collaboration between the groups of Schäfer (Warwick) and Eyice (Birmingham) who have a previous track record of working on the microbiology and ecology of DMS degrading microorganisms. There will be opportunities to collaborate with colleagues from their existing network on other aspects of organic sulfur compound metabolism, bacterial genetics, and environmental omics.
Further details
Applicants are strongly encouraged to contact Hendrik Schäfer ([email protected]) to discuss the project and ask any questions they may have.
To apply to this project:
- You must include a CENTA studentship application form, downloadable from: CENTA Studentship Application Form 2025.
- You must include a CV with the names of at least two referees (preferably three) who can comment on your academic abilities.
- Please submit your application and complete the host institution application process via: https://warwick.ac.uk/fac/sci/lifesci/study/pgr/studentships/nerccenta/ University of Warwick projects will be added here: https://warwick.ac.uk/fac/sci/lifesci/study/pgr/studentships/nerccenta/studentships/ and application guidance is at the bottom of this page. Complete the online application form – selecting course code P-C1PB (Life Sciences PhD); from here you will be taken through to another screen where you can select your desired project. Please enter “NERC studentship” in the Finance section and add Nikki Glover, [email protected] as the scholarship contact. Please also complete the CENTA Studentship Application Form 2025 and submit via email to [email protected]. Please quote CENTA 2025-W20 when completing the application form.
Applications must be submitted by 23:59 GMT on Wednesday 8th January 2025.
Possible timeline
Year 1
Assessment of anaerobic DMS degradation with range of electron acceptors, diversity analysis using 16S rRNA amplicon sequencing, enrichment and beginning of isolation.
Year 2
DNA stable isotope probing of selected samples, metagenomic analysis, characterisation of isolates.
Year 3
Bioinformatic data mining of meta-omics datasets and functional analysis of anaerobic DMS degrading isolate(s).
Further reading
Haaijer, S.C.M., Harhangi, H.R., Meijerink, B.B., Strous, M., Pol, A., Smolders, A.J.P., Verwegen, K., Jetten, M.S.M. and Den Camp, H. (2008). Bacteria associated with iron seeps in a sulfur-rich, neutral pH, freshwater ecosystem. ISME J, 2, 1231-1242
Kappler, U. and Schäfer, H. 2014. Transformations of dimethylsulfide. In: P.M.H. KRONECK, M. E. S. T. (ed.) The Metal-Driven Biogeochemistry of Gaseous Compounds in the Environment. Dordrecht: Springer Science + Business Media B.V.
Lyimo, T.J., Pol, A., Harhangi, H.R., Jetten, M.S.M. and Op Den Camp, H.J.M. (2009). Anaerobic oxidation of dimethylsulfide and methanethiol in mangrove sediments is dominated by sulfate-reducing bacteria. FEMS Microbiol. Ecol., 70, 483-492
Schäfer, H., Myronova, N. and Boden, R. (2010). Microbial degradation of dimethylsulphide and related C1-sulphur compounds: organisms and pathways controlling fluxes of sulphur in the biosphere. J Exp Bot, 61, 315-334
Tanimoto, Y. and Bak, F. (1994). Anaerobic degradation of methylmercaptan and dimethyl sulfide by newly isolated thermophilic sulfate-reducing bacteria. Appl. Environ. Microbiol., 60, 2450-2455
Tebbe, D.A., Gruender, C., Dlugosch, L., Lõhmus, K., Rolfes, S., Könneke, M., Chen, Y., Engelen, B. and Schäfer, H. (2023). Microbial drivers of DMSO reduction and DMS-dependent methanogenesis in saltmarsh sediments. The ISME Journal, 17, 2340-2351
Tsola, S.L., Zhu, Y., Chen, Y., Sanders, I.A., Economou, C.K., Brüchert, V. and Eyice, Ö. (2024). Methanolobus use unspecific methyltransferases to produce methane from dimethylsulphide in Baltic Sea sediments. Microbiome, 12, 3
Visscher, P.T. and Taylor, B.F. (1993). Aerobic and anaerobic degradation of a range of alkyl sulfides by a denitrifying marine bacterium. Appl. Environ. Microbiol., 59, 4083-4089